Abstract
WNT inhibitory factor-1 (WIF1) is an antagonist of the WNT signaling pathway. We investigated the relationship between WIF1 promoter methylation and regulation of the WNT/β-catenin signaling pathway, tumor grade, and survival in patients with astrocytoma. This study included 86 cases of astrocytoma, comprising 20 diffuse astrocytomas and 66 glioblastomas. In addition, 17 temporal lobectomy specimens from patients with epilepsy were included as controls. The ratio of methylated DNA to total methylated and unmethylated DNA (% methylation) was measured by methylation- and unmethylation-specific PCR. Representative tumor tissue was immunostained for WIF1, β-catenin, cyclin D1, c-myc, and isocitrate dehydrogenase 1. Levels of WIF1 promoter methylation, mRNA expression, and protein expression in a glioblastoma cell line were compared before and after demethylation treatment. The mean percent methylation of the WIF1 promoter in astrocytomas was higher than that in control brain tissue. WIF1 protein expression was lower in the tumor group with >5% methylation than in the group with <5% methylation. Cytoplasmic β-catenin staining was more frequently observed in tumors with a low WIF1 protein expression level. Demethylation treatment of a glioblastoma cell line increased WIF1 mRNA and protein expression. Increased WIF1 promoter methylation and decreased WIF1 protein expression were not related to patient survival. In conclusion, WIF1 expression is downregulated by promoter methylation and is an important mechanism of aberrant WNT/β-catenin pathway activation in astrocytoma pathogenesis.
Similar content being viewed by others
Main
Astrocytomas are the most common primary tumors of the central nervous system. Despite aggressive treatment with surgery and subsequent chemoradiotherapy, high-grade astrocytomas show a high rate of recurrence and mortality.1 Understanding the molecular basis of astrocytoma tumorigenesis is necessary for prediction of therapy response, prognosis, and the development of more effective therapies. Recently, several reports showed a relationship between the WNT signaling pathway and the grade and prognosis of astrocytomas.2, 3, 4 Both mRNA and protein expression levels of β-catenin are increased in astrocytoma compared with normal brain tissue.4 They are also higher in high=grade tumors than in low-grade tumors.2, 4 Moreover, cytoplasmic/nuclear β-catenin and cyclin D1 status have been reported to be independent prognostic factors in glioma patients.2
WNT inhibitory factor-1 (WIF1) is an antagonist that inhibits the WNT signaling pathway;5, 6, 7 therefore, functional loss of WIF1 can contribute to tumorigenesis by activation of the WNT pathway. Downregulation of WIF1 mRNA and protein expression levels as a result of WIF1 gene promoter methylation has been reported in human astrocytoma.7, 8 However, the influence of WIF1 promoter methylation and WIF1 protein expression level on components of WNT pathway has not been described in astrocytoma, except for a single study revealing that demethylation of the WIF1 gene promoter and restoration of WIF1 protein expression decreased cytosolic β-catenin protein levels.9 Furthermore, although there is a published study on the relationship between WIF1 promoter methylation and the grade of astrocytomas,7 the prognostic impact of WIF1 promoter methylation and WIF1 protein expression has not been investigated in astrocytomas.
In this study, we primarily investigated the biological and clinical effects of WIF1 gene promoter methylation and WIF1 protein expression with respect to tumor grade and patient survival. We also investigated the role of WIF1 gene promoter methylation as a regulatory mechanism of WIF1, and subsequently, other proteins of WNT pathway such as β-catenin, cyclin D1, and c-myc expression, along with isocitrate dehydrogenase 1 (IDH1).
Materials and methods
Case Selection
We retrieved samples of astrocytoma tumors that were resected or biopsied at Asan Medical Center between 2000 and 2008. A total of 86 samples from 81 patients were available as paraffin-embedded material in large enough quantities to obtain sufficient DNA for molecular studies and construct tissue microarrays for immunohistochemical staining. These samples consisted of 20 diffuse astrocytoms and 66 glioblastomas, according to the WHO classification of tumors of the nervous system.1 Among 66 glioblastomas, 5 were recurrent tumors, and therefore, there were 61 glioblastoma patients. Adjacent nonneoplastic brain tissues were available in 6 diffuse astrocytomas and 12 glioblastomass. For control brain samples, we selected 17 temporal lobectomy specimens from patients with epilepsy. We also collected data on patient age, sex, tumor size, residual tumor after resection, and survival.
Cell Culture and In Vitro Demethylation Treatment
The human glioblastoma cell line U251, listed among the NCI-60 cell lines (Jackson Laboratory, Sacramento, CA, USA), was used in this study. Cells were grown as monolayers in RPMI-1640 medium supplemented with 10% heat inactivated fetal bovine serum and 1% penicillin–streptomycin solution (Gibco, Carlsbad, CA, USA). Cultures were maintained at 37 °C in a humidified atmosphere containing 5% CO2 with a change of culture medium every 2–3 days. For demethylation treatment, cells were seeded in 100-mm cell culture dishes (Falcon, Franklin Lakes, NJ, USA) for 24 h before treatment with 10 μM 5-aza-2′-deoxycytidine (5-AZA-dC, Sigma Aldrich, St Louis, MO, USA) for 3 days. Fresh drug was added every 24 h.
Genomic DNA Extraction
Genomic DNA extraction from paraffin-embedded tissue was performed with QIAamp DNA FFPE Tissue Kit (QIAgen, Hilden, Germany). Genomic DNA was extracted from U251 cells using Labopass GenomicTM isolation Kit (COSMO, Genetech, Seoul, Korea) according to the manufacturer’s instructions. Before methylation- and unmethylation-specific PCR, bisulfite modification was performed with 2 μg genomic DNA using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA) to obtain a final elution volume of 10 μl. Treatment of DNA with bisulfite resulted in the conversion of unmethylated cytosine into uracil. In contrast, 5-methyl cytosine (mC) was not converted and thus remained as cytosine.
Methylation- and Unmethylation-Specific PCR
The promoter region of the WIF1 gene was identified using the transcriptional regulatory element database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm; Accession number, 10003). The PCR primers targeted CpG islands of the WIF1 promoter region. The sequences of the methylation-specific and unmethylation-specific reverse primers were 5′-ACGCGAACGAAATACGAACG-3′ and 5′-CCCACAAAACCTAAACAACCA-3′, respectively. The forward primer (5′-ATTGGGYGTATTGTATTGTGAATG-3′) was designed to anneal equally well to methylated and unmethylated DNA. For methylation- and unmethylation-specific PCR using genomic DNA from paraffin-embedded tissue, 1.5 μl each of forward primer and methylation-specific reverse primer at 10 pmol concentration were mixed with 1 μl unmethylation-specific primer at the same concentration for effective detection of methylated DNA. For PCR with genomic DNA from the glioblastoma cell line, we used 1.5 μl forward primer, 1 μl methylation-specific reverse primer, and 1.5 μl unmethylation-specific reverse primer. The three primers were mixed in one tube together with 2 μl bisulfate-converted DNA to compare the proportions of methylated and unmethylated DNA in the WIF1 promoter. PCR was performed using Blend Taq-Plus polymerase (TOYOBO, Osaka, Japan) and the following cycling conditions: an initial denaturation step at 94 °C for 5 min followed by 20 cycles of 15 s at 94 °C; 30 s at the annealing temperature, which started at 60 °C and decreased by 0.5 °C in each cycle; and 30 s at 72 °C. This was followed by an additional 20 cycles of 94 °C for 15 s, 56 °C for 30 s, and 72 °C for 30 s, with a final incubation at 72 °C for 5 min. The amplified products were analyzed by electrophoresis on 3% agarose gels containing ethidium bromide and visualized under ultraviolet light.
Quantification of WIF1 Promoter Methylation
Each sample yielded PCR products of 134 base pairs and 104 base pairs, corresponding to methylated and unmethylated template, respectively (Figure 1). The intensity of each band was measured using AlphaEaseFC software and given as the relative percentage of the combined intensity of the methylation band and unmethylation band, which was taken as 100%. For quantification of the amount of methylated DNA in proportion to total DNA, we constructed a standard curve using control DNA samples prepared by mixing totally methylated DNA and totally unmethylated control DNA (EpiTect PCR Control DNA set, bisulfate-converted; QIAgen) in proportions of 0, 5, 10, 20, 50, 80, 90, 95, and 100% methylated DNA (Supplementary Figures 1a and b). Methylation- and unmethylation-specific PCR was performed using the control DNA and the two primer mixes described above for samples from paraffin-embedded tissues and the U251 cell line. Fitting of the standard curves resulted in the functional formula y=2.598e0.035x for the primer ratio used for PCR of DNA from paraffin-embedded tissue (Supplementary Figure 1c) and y=2.671e0.034x for the primer ratio used for PCR of DNA from the U251 cell line (Supplementary Figure 1d), where x represents the relative intensity of the methylation band, and y represents the proportion of methylated DNA in total DNA (% methylation).
Methylation- and unmethylation-specific PCR bands. Methylation-specific bands are evident at 134 bp and unmethylation-specific bands at 104 bp. Unmethylation bands were observed in all samples. Methylation bands were frequently observed in glioblastoma (GB) samples, but less frequently observed in diffuse astrocytoma (DA), adjacent brain tissue (AN), and control brain tissue (N). DW, distilled water control; UC, 100% unmethylated control DNA; MC, 100% methylated control DNA.
Measurement of WIF1 mRNA in Glioblastoma Cell Line
Total RNA was extracted from U251 cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and quantified using NanoDrop Technologies (Wilmington, DE, USA). Complementary DNA was synthesized from 1 μg of extracted total RNA using oligo-dT primers and a SuperScript III reverse transcriptase kit (Invitrogen). Forward and reverse primers used to amplify the WIF1 cDNA gene were 5′-CCGAAATGGAGGCTTTTGTA-3′ and 5′-TGGTTGAGCAGTTTGCTTTG-3′, respectively. Each 20 μl reaction contained 0.5 units of Blend Taq-Plus-polymerase, 1 × PCR buffer for Blend Taq, 1 pmol of each primer, and 0.2 mM dNTP. PCR cycling conditions were initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 56 °C for 40 s, and 72 °C for 40 s, with a final extension at 72 °C for 10 min. The amplified products were analyzed by electrophoresis on 2% agarose gels containing ethidium bromide and visualized under ultraviolet light. The intensity of each band was measured using AlphaEaseFC software and normalized to β-actin.
Construction of Tissue Microarray Blocks and Immunohistochemistry
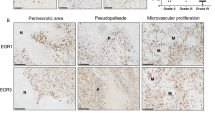
For immunohistochemistry, tissue microarrays were constructed with one representative core of 2-mm diameter from formalin-fixed, paraffin-embedded samples. Sections of 4 μm thickness were obtained from tissue microarrays with a microtome, transferred onto adhesive slides, and dried at 62 °C for 30 min. Immunohistochemical staining with antibody specific for WIF1 (1:200; R&D Systems, Minneapolis, MN, USA) was performed using Dako EnVision+system-HRP (Dako, Glostrup, Denmark). Immunohistochemical staining with antibody against β-catenin (1:2000; Zymed, San Francisco, CA, US), cyclin D1 (1:100; Neomarkers, San Francisco, CA, USA), c-myc (1:50; Novocastra, Notting Hill, UK), and IDH1 (anti-IDH1R132H, 1:50; Dianova, Hamburg, Germany) was performed using a Benchmark automatic immunostaining device (Ventana Medical System, Tucson, AZ, USA). WIF1 expression was considered positive when staining was present in the cytoplasm. Staining was scored for intensity (0 (negative), 1 (weak), 2 (strong)) and for percentage of positive tumor cells among all tumor cells (0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), 4 (76–100%)). For cyclin D1, nuclear staining intensity and percentage of positive tumor cells were assigned as follows: intensity score of 0 (negative), 1 (weak), 2 (intermediate), or 3 (strong); and percentage score of 0 (0%), 1 (1–5%), 2 (10–30%), or 3 (40–100%). The final immunohistochemical score for WIF1 and cyclin D1 was obtained by multiplication of the intensity and percentage scores. C-myc was considered positive when expressed in cytoplasm, as per the manufacturer’s protocol. Staining for c-myc was homogenous in all astrocytomas, and therefore the immunohistochemical score only reflected intensity, scored as 0 (negative), 1 (weak), 2 (intermediate), or 3 (strong). Staining patterns of β-catenin were divided into membranous, cytoplasmic, and nuclear staining. Combined membrane and cytoplasmic and combined cytoplasmic and nuclear pattern were also described (Figure 2). Adjacent nonneoplastic brain tissue and control brain tissue were not amenable to immunohistochemical scoring because nonneoplastic glial cells have very small amounts of cytoplasm and mainly consist of neuropils.
Immunohistochemical staining. (a, b, c) Negative, weak, and strong cytoplasmic WIF1 protein expression. (d, e) Membranous and cytoplasmic β-catenin staining. (f) β-Catenin staining in both membrane and cytoplasm. (g, h, i) Weak, intermediate, and strong nuclear cyclin D1 protein expression. (j, k, l) Weak, intermediate, and strong cytoplasmic c-myc expression.
Immunohistochemical Staining and Immunofluorescence Assay for WIF1 in Glioblastoma Cell Line
Expression of WIF1 protein in glioblastoma cells was measured by immunohistochemical staining and immunofluorescence assay before and after treatment with 5-AZA-dC. The cells were seeded in a 4-well slide chamber (Lab-Tek) and fixed with 4% paraformaldehyde for 10 min. Before immunostaining, antigen was retrieved by incubation in 10 mM sodium citrate (pH 6.0) for 10 min. The cells were incubated in peroxidase blocking buffer (0.3% H2O2 in methanol) to block activity of endogenous peroxides, and then with 1% goat serum and 1% bovine serum albumin in 0.1 M PBS to block nonspecific antibody binding. Cells were treated with antibody against human WIF1 (1:100; R&D Systems) at room temperature for 1 h. Substrate-diaminobenzidine (DAB) (Envision DAB+ Kit, Dako) was added and the samples were counterstained with hematoxylin (Mayer’s hematoxylin; Dako). The percentage of WIF1-positive cells was determined by counting a total of 200 cells and scoring positively stained cells.
For the immunofluorescence assay, cells were fixed with 4% PFA in 0.1 M PBS (pH 7.4) for 10 min. The fixed cells were incubated in 0.1% Triton-X 100 in 0.01 M sodium citrate buffer for 15 min for antigen retrieval, and then washed with PBS washing buffer (0.05% Tween-20 in 0.1 M PBS). Nonspecific antigens were removed using 1% BSA/1% NGS in 0.1 M PBS solution. The cells were incubated with anti-WIF1 antibody for 1 h at room temperature. WIF1 antibody was detected by incubation with goat anti-mouse IgG-conjugated Alexa 555 secondary antibody (Invitrogen) for 1 h. Each sample was counterstained with DAPI (Invitrogen) and mounted with flourescence mounting solution.
Statistical Analyses
Comparison of patient age between different groups was performed using Student’s t-test. Variables that did not have a normal distribution, such as % methylation and WIF1 immunohistochemical score, were analyzed by Mann–Whitney U-test. To adjust for the age effect on % methylation, we used simple and multiple linear regression models after transformation of % methylation to log(% methylation+1) to give a normal distribution. The correlation between promoter methylation, protein expression of WIF1, β-catenin, cyclin D1, c-myc, IDH1, and tumor grade was analyzed using Pearson’s χ2 test or Fisher’s exact test, with a P-value <0.05 considered to be statistically significant. For measurement of WIF1 promoter methylation, mRNA, and protein expression level in the U251 cell line, each experiment was performed three times to provide a mean value, and means were compared by Mann–Whitney U-test. Univariate and multivariate survival analyses were performed using the Cox regression hazard model. All statistical analyses were performed with SPSS version 15.0.
Results
Clinical Data
Among 66 glioblastomas, 5 were recurrent tumor, the patients with which were included in glioblastoma groups. Therefore, 61 patients with glioblastoma, 20 patients with diffuse astrocytoma, and 17 patients who underwent temporal lobectomy were included in the analysis of the age, sex, and survival of patients. Male-to-female ratios were 1.1:1 for the glioblastoma group, 4:1 for the diffuse astrocytoma group, and 1.8:1 for the temporal lobectomy group. Mean patient ages were 45.3 years (10–72 years) for glioblastoma, 38.0 years (24–69 years) for diffuse astrocytoma group, and 34.8 years (27–46 years) for the temporal lobectomy group. The mean age of patients with tumor (diffuse astrocytoma and glioblastoma) was 43.5 years (10–72 years) and was significantly higher than that of patients in temporal lobectomy group providing control brain tissue. The mean age of patients with glioblastoma was greater than that of patients with diffuse astrocytoma (P<0.001). The mean age of patients whose samples contained nonneoplastic brain tissue adjacent to the tumors was 42.8 years (10–65 years), and was significantly greater than that of the temporal lobectomy group (P=0.031). The mean age of patients with glioblastoma or diffuse astrocytoma was significantly higher than that of patients in the temporal lobectomy group (P<0.001).
Among 66 glioblastoma, 52 tumors were subjected to postoperative adjuvant radiotherapy (22, 33.3%), chemotherapy (11, 16.7%), and chemoradiotherapy (19, 28.8%). No adjuvant therapy was performed in 8 (12.1%) glioblastoma. Because 6 patients (9.1%) with glioblastoma were transferred to other hospital after operation, we could not figure out whether adjuvant therapy was done or not. For diffuse astrocytoma, 12 (60.0%) tumors were subjected to radiotherapy, and 1 (5.0%) tumor was subjected to chemotherapy. Adjuvant therapy was not performed in 7 (35.0%) diffuse astrocytomas. Chemotherapeutic agent was tenozolomide in all cases involving chemotherapy.
WIF1 Promoter Methylation Level of Glioblastoma, Diffuse Astrocytoma, Nonneoplastic Brain Tissue Adjacent to Tumor, and Control Brain Tissue
The mean % methylation of the WIF1 promoter in tumors (diffuse astrocytoma and glioblastoma) was 5.2%; this was significantly higher than that in control brain tissues from the temporal lobectomy group (mean 0.2%; P=0.002). Among tumor groups, the mean % methylation was not significantly different between the diffuse astrocytoma group (1.7%) and the glioblastoma group (6.2%; P=0.182; Figure 3). Promoter methylation is known to increase with age. Therefore, to confirm the difference in % methylation between control brain tissues and tumors, adjustment for the age factor was needed because patient age was significantly different between these groups.10 On multivariate analysis with a linear regression model, the higher % methylation in tumor than in control brain tissue remained statistically significant, independent of patient age (P=0.015; Table 1).
Although the methylation band was detected in three samples of control brain tissue from the temporal lobectomy group (17.3%), all control tissues had <5% methylation, whereas 20 of the tumors (23.3%) showed >5% methylation (P=0.038). We set 5% methylation as the cutoff value for dividing the tumor group because up to 5% methylation could be observed in control brain tissue and therefore might reflect aging, the disease process of epilepsy, or false-positive methylation caused by incomplete bisulfate modification reaction or nonspecific amplification. As described in Table 2, the proportion of patients with >5% methylation was higher for glioblastoma (17 of 66, 25.8%) than diffuse astrocytoma (3 of 20, 15.0%) but this was not significant (P=0.382), consistent with the results for mean % methylation.
Adjacent nonneoplastic brain tissue was available in 6 diffuse astrocytoma cases and 12 glioblastoma cases. Of the 18 adjacent brain tissue samples, 9 (50.0%) yielded a methylation band on methylation- and unmethylation-specific PCR, and 1 of them (5.6%) showed >10% methylation (19.2). The mean % methylation of adjacent brain tissue was 2.3%, and was significantly higher than 0.2% of control brain tissue from the TL group (P=0.041; Figure 3). However, when we considered the younger mean age of patients in the temporal lobectomy group compared with the group with adjacent brain tissues, multivariate analysis to adjust for the age effect revealed only a marginally significant difference between % methylation of the two groups (P=0.073; Table 1). There was a positive correlation between % methylation of the tumor and the respective adjacent nonneoplastic tissue (Spearman’s correlation coefficient 0.564, P=0.015).
Immunohistochemical Analysis of Paraffin-Embedded Tumor Tissue to Investigate the Correlation Between WIF1 and Other Proteins in the WNT/β-Catenin Pathway
The results of immunohistochemical staining for WIF1, β-catenin, cyclin D1, and c-myc are summarized in Tables 2 and 3. Four β-catenin staining patterns were identified. Among 86 astrocytomas, 51 tumors (59.3%) showed membranous pattern, 6 tumors (7.0%) showed membranous and cytoplasmic patterns, 23 tumors (26.7%) had cytoplasmic pattern, and 6 tumors (7.0%) had cytoplasmic and nuclear patterns. In all of the 6 tumors (1 diffuse astrocytoma and 5 glioblastomas) with combined membranous and cytoplasmic β-catenin staining patterns, the β-catenin positivity was stronger in membrane than in cytoplasm. The 6 tumors (2 diffuse astrocytomas and 4 glioblastomas) with nuclear β-catenin positivity had weak to strong cytoplasmic positivity. Therefore, for statistical analysis, the locations of β-catenin staining were divided into two groups: membrane or membrane/cytoplasm and cytoplasm or cytoplasm/nucleus. Mean proportion of WIF1-positive cells was lower in glioblastomas than in diffuse astrocytomas (P=0.003; Table 2). More than 50% of tumor cells were positive for WIF1 in 75% (15 of 20) of diffuse astrocytomas, compared with 33.3% (22 of 66) of glioblastomas (P=0.002). Similarly, 75% (15 of 20) of diffuse astrocytomas had a WIF1 immunohistochemical score of 3–8, compared with 36.4% (24 of 66) of glioblastomas (P=0.004). A cytoplasmic and/or nuclear β-catenin staining pattern was more frequently observed in glioblastomas (39.4%) than in diffuse astrocytomas (15.0%; P=0.059). More glioblastomas than diffuse astrocytomas had a cyclin D1 immunohistochemical score of 4–9 (P=0.020) and the intensity of c-myc staining was stronger in glioblastomas than in diffuse astrocytomas (P=0.002). The proportion of IDH1-positive tumors was slightly higher in diffuse astrocytomas (20.0%) than in glioblastomas (13.6%), but statistical significance was not reached (P=0.489).
Of the 49 tumors containing ≤50% WIF1-positive cells, 24 (49.0%) had cytoplasmic or cytoplasmic/nuclear β-catenin staining pattern, a significantly higher rate than in tumors containing >50% WIF1 positive cells (5 of 37 tumors, 13.5%; P=0.001; Table 3). When compared according to WIF1 immunohistochemical score, tumors with cytoplasmic or cytoplasmic/nuclear β-catenin staining pattern were observed in 22 of 47 (46.8%) tumors with immunohistochemical score 0–2, and in 7 of 39 (17.9%) tumors with immunohistochemical score 3–8 (P=0.006). There was no significant relationship between WIF1 protein expression level and expression of cyclin D1, c-myc, or IDH1.
Correlation of WIF1 Promoter Methylation Level and Immunohistochemical Results in Paraffin-Embedded Tumor Tissue
Comparison of immunohistochemical results between the tumor group with ≤5% methylation and the tumor group with >5% methylation is described in Table 4. The mean percentage of tumor cells that were WIF1 positive was lower in the group with >5% methylation than in the group with ≤5% methylation (P=0.001). In the group with >5% methylation, only 3 of 20 tumors (15.0%) contained >50% cells positive for WIF1 and had a WIF1 immunohistochemical score of ≥3; this was strikingly different from the group with ≤5% methylation in which more than half of the tumor cells were positive for WIF1 in 81.5% of cases (P=0.004) and 54.5% showed a WIF1 immunohistochemical score between 3 and 8 (P=0.002). More cases showed cytoplasmic or cytoplasmic/nuclear β-catenin staining pattern in the >5% methylation group than in the ≤5% methylation group, but this difference was not statistically significant (P=0.106). Cyclin D1 immunohistochemical score and c-myc immunohistochemical intensity were slightly higher in the >5% methylation group than in the ≤5% methylation group, but did not reach statistical significance (P=0.241 and 0.251, respectively).
Comparison of WIF1 Promoter Methylation Level and mRNA and Protein Expression of WIF1 in Glioblastoma Cell Line Before and After Demethylation Treatment
In the U251 cell line, the mean % methylation of the WIF1 gene promoter was 65.4% (±8.7 s.d.) without exposure to 5-AZA-dC and decreased to 42.9±7.1 after 5-AZA-dC treatment (P=0.029; Supplementary Figure 2a). After demethylation treatment with 5-AZA-dC, the level of WIF1 mRNA significantly increased 1.9±0.6-fold (P=0.029; Supplementary Figure 2b). On immunohistochemical staining, the percentage of WIF1-positive cells was 20.0±1.0% before 5-AZA-dC treatment and significantly increased to 36.3±2.75% with treatment (P=0.029; Supplementary Figure 2c). In the immunofluorescence assay, more WIF1-positive cells were observed and WIF1 immunostaining was stronger among cells exposed to 5-AZA-dC than among cells without 5-AZA-dC treatment (Supplementary Figures 2d and e).
Survival Analyses
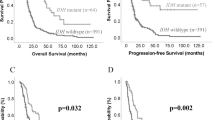
Among the 20 patients with diffuse astrocytoma, 3 (15.0%) patients died, whereas the mortality rate of patients with glioblastoma was 83.6% (51 of 61). Overall patient survival was 77.6, 56.2, 39.2, 35.5, and 28.9% for 1, 2, 3, 4, and 5 years. For diffuse astrocytoma, 3-year survival was 100%, and 4- and 5-year survival was 92.3 and 82.1%, respectively. For glioblastoma, the 1, 2, 3, 4, and 5-year survival was 71.8, 41.5, 20.1, 20.1, and 12.6%, respectively. Mean survival was 81.9 months for diffuse astrocytoma and 28.6 months for glioblastoma. The higher survival rate of patients with diffuse astrocytoma than that of patients with glioblastoma was statistically significant (P<0.001; Figure 4a). On univariate analyses, other factors related to poor survival were increased age (P=0.001) and presence of residual tumor after resection (P=0.065; Table 5). Percent methylation, presented as either a continuous variable or a categorical variable with a cutoff of 5% methylation, had no significant effect on patient survival (Table 5 and Figure 4b). WIF1 positivity in ≤50% of the tumor cells and WIF1 immunohistochemical score of ≤2 were related to poorer patient survival on univariate analysis (P=0.010 and 0.015, respectively; Figures 4c and d). Tumor size, β-catenin, cyclin D1, c-myc, IDH1 staining patterns, and postoperative treatment were not related to patient survival (Table 5). For multivariate analyses, we could select maximum three variables, because only 27 patients survived. Each of WIF1-positive cell percentage and WIF1 immunohistochemical score was entered with tumor grade and patient age into Cox regression hazard model for multivariate analyses (Table 6) that resulted in no significant effect of WIF1-positive cell percentage and WIF1 immumohistochemical score on patient survival (P=0.241 and P=0.258, respectively). Tumor grade and increased patient age were independent significant factors for poorer patient survival (P<0.001, both).
When a set consisting only of 61 glioblastomas were separately analyzed for patient survival (Table 7), increased patient age and presence of residual tumor were adverse prognostic factors (P=0.001 and P=0.008, respectively), but WIF1-positive cell percentage and WIF1 immunohistochemical were not (P=0.367 and P=0.543, respectively), neither were % methylation of WIF1 promoter, tumor size, β-catenin, cyclin D1, c-myc, IDH1 staining patterns, and postoperative treatment. Multivariate analysis was not eligible because only 10 patients survived.
Discussion
WIF1 is known to be downregulated in many malignant tumors such as hepatocellular carcinoma,11, 12 nasopharyngeal carcinoma,13, 14 lung cancer,9, 15 colon cancer,14 renal cell carcinoma,16 esophageal adenocarcinoma,17 melanoma,18 and bladder cancer.19 The main mechanism of WIF1 inactivation is aberrant promoter methylation, as demonstrated by the inverse correlation between WIF1 promoter methylation and expression of WIF1 mRNA or protein8, 11, 12, 19, 20, 21, 22, 23, 24 and restoration of WIF1 expression by promoter demethylation treatment.9, 20, 22, 23 Yang et al7 first described WIF1 gene promoter hypermethylation in human astrocytoma and showed that this methylation was associated with decreased Wif1 mRNA and protein expression. In their report, the frequency of aberrant promoter methylation was higher in astrocytoma than in normal brain tissue, where no aberrant methylation was found. In our series, the level of WIF1 gene promoter methylation, described as % methylation, was higher in astrocytoma than in control brain tissue from epilepsy patients, consistent with results from previous studies of astrocytoma and tumors of other organs. However, patient age should be considered before drawing any conclusions, because a recent study by Hernandez et al10 revealed a positive correlation between age and DNA methylation level in human brain tissue. In our study, the higher level of WIF1 promoter methylation in astrocytoma was statistically significant independent of the age difference between the astrocytoma group and the control temporal lobectomy group, as proved by multivariate analysis using a linear regression model. This finding strongly suggests that WIF1 promoter methylation plays a major role in the pathophysiology of astrocytoma.
Several authors support the theory that promoter methylation occurs in the early stage of carcinogenesis based on the observation that genes of nonneoplastic tissue from organs containing tumor are more frequently methylated than those of tissue from patients without tumor.14, 17, 24 However, studies on the methylation status of nonneoplastic tissue adjacent to tumors of various organs revealed controversial results. It has been reported that WIF1 promoter hypermethylation is more frequent in adjacent normal colorectal mucosa from cancer patients compared with normal mucosa from patients without tumor.24 However, a study on hepatocellular carcinoma revealed no significant difference in WIF1 promoter methylation frequency between adjacent normal tissue and normal control tissue.11 Our results showed a higher methylation level in nonneoplastic brain tissue adjacent to astrocytoma than in control tissue from epileptic brain. Astrocytomas are not amenable to wide excision because of high morbidity after large resection of brain tissue, and are frequently infiltrative in microscopic level. Therefore, adjacent brain tissue in this study might contain tumor cells from the first place, and might influence on increased promoter methylation level. Other explanation is that the younger mean age of patients in the control group seemed to influence this result because the effect was canceled by multivariate analysis adjusting for the age factor. Larger numbers of tumor cases with adjacent brain tissue and age-matched control brain tissue are needed for more reliable investigation of methylation status in adjacent brain tissue.
An inverse correlation between promoter methylation of the WIF1 gene and expression of WIF1 mRNA or protein8, 11, 12, 19, 20, 22, 23, 24 and restoration of WIF1 expression by promoter demethylation treatment9, 20, 22, 23 have been shown in many malignant tumors including astrocytoma.7, 8 Our study also revealed an inverse correlation between promoter methylation and WIF1 protein expression, and an increase in WIF1 mRNA and protein expression level following demethylation treatment, confirming that promoter methylation is a major inactivation mechanism of the WIF1 gene.
Our results regarding tumor grade showed that expression of WIF1, β-catenin, cyclin D1, and c-myc were all associated with tumor grade, but WIF1 promoter methylation was not. The relationship between promoter methylation of the WIF1 gene and tumor grade is controversial. A study on colon cancer showed that WIF1 promoter methylation was not related to tumor grade,24 in contrast to the result of Yang et al7 who reported a higher frequency of aberrant methylation of the WIF1 promoter in glioblastomas than in low-grade gliomas. In our study, both the mean % methylation and the proportion of tumors with >5% methylation were higher in glioblastoma than in diffuse astrocytomas, although statistical significance was not reached. Further study with more astrocytoma cases is warranted to clarify this issue. On the other hand, the reduced level of WIF1 protein expression, more frequent cytoplasmic or cytoplasmic/nuclear β-catenin staining pattern, and increased cyclin D1 and c-myc expression in glioblastomas relative to diffuse astrocytomas were consistent with results of other studies.2, 4, 7, 25 These findings suggest that WIF1 promoter methylation is a common process that occurs relatively upstream in tumorigenesis, and that the accumulation of additional downstream alterations in the WNT/β-catenin pathway is related to the aggressiveness of astrocytoma.
Regarding β-catenin immunohistochemistry, we lumped four β-catenin staining patterns into two groups for statistical analysis. The one was membranous or membranous/cytoplasmic pattern, and the other was cytoplasmic and cytoplasmic/nuclear pattern. β-Catenin plays a pivotal role in canonical Wnt signaling pathway and cell-to-cell adhesion by linking cadherins and actin cytoskeleton. Therefore, the β-catenin expression is membranous in normal and nonneoplastic cells. In contrast, as presented in many studies, cytoplasmic accumulation is observed in a significantly higher proportion of malignant tumors than of normal or nonneoplastic tissue.4, 26, 27, 28 This method of grouping actually revealed significant correlation between β-catenin and other markers such as cyclin D1, c-myc, APC, E2F1, p53, and MDM2, and tumor grade and patient survival.4, 26, 27 The β-catenin expression in the cytoplasm and/or nucleus could be considered to be an indication of its aberrant expression. The association between decreased WIF1 expression and cytoplasmic or cytoplasmic/nuclear staining of β-catenin is similar to the results of a study by Gao et al,9 in which promoter demethylation treatment increased WIF1 expression and decreased the cytosolic β-catenin level. However, expression of cyclin D1 or c-myc was not related to the expression level of WIF1 or to WIF1 promoter methylation, although the proportion of cells with intermediate or strong c-myc immunohistochemical intensity was slightly higher in tumors with decreased WIF1 immunohistochemical expression. This contrasts with the study of bladder cancer by Urakami et al,19 in which expression of cyclin D1 and c-myc was higher in tumors with low WIF1 mRNA expression. It is apparent that β-catenin is regulated by WIF1 through WNT signaling. However, although WIF1 may play a role in the regulation of cyclin D1 and c-myc, we should also consider the involvement of other mechanisms.
The mutation of IDH1 in glioma is known to be associated with young age, a secondary-type GBM, and increased overall survival.29, 30 In our study, the proportion of IDH1-positive tumor was lower than reported,29, 30 and was not significantly different between diffuse astrocytoma and glioblastoma, between low and high WIF1 positivity group, either between <5% methylation or >5% methylation group. IDH1-positive tumors showed longer patient survival, although statistical significance was not reached. Although the oncogenic mechanism of IDH1 mutation remains largely unknown, mutant IDH1 seems to induce global DNA methylation.31 Therefore, WIF1 promoter methylation might be affected by IDH1 mutation, but further study with larger numbers of cases is necessary to confirm.
The role of promoter methylation of the WIF1 gene as a prognostic factor is still controversial. Promoter methylation of WIF1 was reported to have no association with patient survival in hepatocellular carcinoma,11, 12 but a relationship with TNM stage and age was demonstrated in nasopharyngeal carcinoma.12 In stage IA non-small cell lung cancer, WIF1 promoter methylation is an independent prognostic factor in relapse-free survival and patient survival.15 The association between WIF1 promoter methylation or WIF1 protein expression level and patient survival has not previously been investigated in astrocytoma. In this study of astrocytoma, % methylation of the WIF1 gene promoter showed no association with patient survival. Patients with decreased WIF1 protein expression in their tumors showed shorter survival than patients with high WIF1 protein expression on univariate analysis, but the statistical significance was diminished by multivariate analysis with tumor grade and patient age. The univariate survival analysis with a separate set consisting only of glioblastomas showed similar results. It seems that astrocytomas with decreased WIF1 protein expression show worse prognosis because tumors with low WIF1 protein expression are more frequently high-grade tumors. A recent study8 with glioblastoma cell line revealed that WIF1 overexpression induced a senescence-like phenotype, inhibited cell proliferation and anchorage-independent growth in vitro, and abolished tumorigenicity in vivo, suggesting that WIF1 can be a therapeutic target in the treatment of astrocytoma. Therefore, the independent prognostic effect of WIF1 promoter methylation and WIF1 protein expression is yet to be revealed with larger numbers of tumor samples.
In conclusion, promoter methylation of the WIF1 gene is increased in astrocytoma compared with control brain tissue, although the level of methylation is not associated with tumor grade, suggesting that WIF1 promoter methylation is associated with initiation of astrocytoma. In contrast, accumulation of additional downstream alterations in the WNT/β-catenin pathway, such as expression of β-catenin, cyclin D1, and c-myc, seems to be associated with more aggressive tumor behavior. Promoter methylation is a major mechanism of WIF1 gene inactivation, and decreased WIF1 protein expression is associated with increased accumulation of cytoplasmic or cytoplasmic/nuclear β-catenin, supporting a role of an intimately linked mechanism involving WIF1 promoter methylation, WIF1 expression, and the WNT/β-catenin pathway in astrocytoma tumorigenesis. WIF1 promoter methylation and WIF1 protein expression are not related to patient survival, but further studies with larger numbers of materials are needed to evaluate their prognostic effect.
References
David N, Louis HO, Otmar D et al(eds) WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer: Lyon, 2007, pp 25–49.
Liu C, Tu Y, Sun X et al. Wnt/beta-Catenin pathway in human glioma: expression pattern and clinical/prognostic correlations. Clin Exp Med 2011;11:105–112.
Pu P, Zhang Z, Kang C et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther 2009;16:351–361.
Sareddy GR, Panigrahi M, Challa S et al. Activation of Wnt/beta-catenin/Tcf signaling pathway in human astrocytomas. Neurochem Int 2009;55:307–317.
Hsieh JC, Kodjabachian L, Rebbert ML et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 1999;398:431–436.
Kawano Y, Kypta R . Secreted antagonists of the Wnt signalling pathway. J Cell Sci 2003;116 (Pt 13):2627–2634.
Yang Z, Wang Y, Fang J et al. Expression and aberrant promoter methylation of Wnt inhibitory factor-1 in human astrocytomas. J Exp Clin Cancer Res 2010;29:26.
Lambiv WL, Vassallo I, Delorenzi M et al. The Wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol 2011;13:736–747.
Gao Z, Xu Z, Hung MS et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res 2009;29:2025–2030.
Hernandez DG, Nalls MA, Gibbs JR et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet 2011;20:1164–1172.
Ding Z, Qian YB, Zhu LX et al. Promoter methylation and mRNA expression of DKK-3 and WIF-1 in hepatocellular carcinoma. World J Gastroenterol 2009;15:2595–2601.
Huang L, Li MX, Wang L et al. Prognostic value of Wnt inhibitory factor-1 expression in hepatocellular carcinoma that is independent of gene methylation. Tumour Biol 2011;32:233–240.
Fendri A, Khabir A, Hadri-Guiga B et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter is common and occurs in advanced stage of Tunisian nasopharyngeal carcinoma. Cancer Invest 2010;28:896–903.
Lee BB, Lee EJ, Jung EH et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res 2009;15:6185–6191.
Yoshino M, Suzuki M, Tian L et al. Promoter hypermethylation of the p16 and Wif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol 2009;35:1201–1209.
Kawakami K, Hirata H, Yamamura S et al. Functional significance of Wnt inhibitory factor-1 gene in kidney cancer. Cancer Res 2009;69:8603–8610.
Clement G, Guilleret I, He B et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter occurs early in the carcinogenesis of Barrett’s esophagus. Cancer Sci 2008;99:46–53.
Lin YC, You L, Xu Z et al. Wnt inhibitory factor-1 gene transfer inhibits melanoma cell growth. Hum Gene Ther 2007;18:379–386.
Urakami S, Shiina H, Enokida H et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res 2006;12:383–391.
Batra S, Shi Y, Kuchenbecker KM et al. Wnt inhibitory factor-1, a Wnt antagonist, is silenced by promoter hypermethylation in malignant pleural mesothelioma. Biochem Biophys Res Commun 2006;342:1228–1232.
Huang L, Li MX, Wang L et al. Prognostic value of Wnt inhibitory factor-1 expression in hepatocellular carcinoma that is independent of gene methylation. Tumour Biol 2010;32:233–240.
Lin YC, You L, Xu Z et al. Wnt signaling activation and WIF-1 silencing in nasopharyngeal cancer cell lines. Biochem Biophys Res Commun 2006;341:635–640.
Mazieres J, He B, You L et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res 2004;64:4717–4720.
Qi J, Zhu YQ, Luo J et al. [The role of secreted Wnt-antagonist genes hypermethylation in early detection of colorectal tumor]. Zhonghua Yi Xue Za Zhi 2007;87:1954–1957.
Sareddy GR, Challa S, Panigrahi M et al. Wnt/beta-catenin/Tcf signaling pathway activation in malignant progression of rat gliomas induced by transplacental N-ethyl-N-nitrosourea exposure. Neurochem Res 2009;34:1278–1288.
Liu C, Tu Y, Sun X et al. Wnt/beta-Catenin pathway in human glioma: expression pattern and clinical/prognostic correlations. Clin Exp Med 2010;11:105–112.
Kotsinas A, Evangelou K, Zacharatos P et al. Proliferation, but not apoptosis, is associated with distinct β-catenin expression patterns in non-small-cell lung carcinomas: relationship with adenomatous polyposis coli and G1-to S-phase cell-cycle regulators. Am J Pathol 2002;161:1619–1634.
Huang CL, Liu D, Ishikawa S et al. Wnt1 overexpression promotes tumour progression in non-small cell lung cancer. Eur J Cancer 2008;44:2680–2688.
Guo C, Pirozzi CJ, Lopez GY et al. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr Opin Neurol 2011;24:648–652.
Camelo-Piragua S, Jansen M, Ganguly A et al. A sensitive and specific diagnostic panel to distinguish diffuse astrocytoma from astrocytosis: chromosome 7 gain with mutant isocitrate dehydrogenase 1 and p53. J Neuropathol Exp Neurol 2011;70:110–115.
Figueroa ME, Abdel-Wahab O, Lu C et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18:553–567.
Acknowledgements
We appreciate Wha Jung Kim and Min Ju Kim for their assistance in statistical analyses. This work was supported by Grants (2010-0028034: the Basic Science Research Program; NRF-2010-R13-0029521) from the National Research Foundation of Korea and the Asan Institute for Life Science (2012-526).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Kim, S., Kwak, J., Nam, H. et al. Promoter methylation of WNT inhibitory factor-1 and expression pattern of WNT/β-catenin pathway in human astrocytoma: pathologic and prognostic correlations. Mod Pathol 26, 626–639 (2013). https://doi.org/10.1038/modpathol.2012.215
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.215
Keywords
This article is cited by
-
Cellular retinol binding protein-1 inhibits cancer stemness via upregulating WIF1 to suppress Wnt/β-catenin pathway in hepatocellular carcinoma
BMC Cancer (2021)
-
Omega-3 PUFA Alters the Expression Level but Not the Methylation Pattern of the WIF1 Gene Promoter in a Pancreatic Cancer Cell Line (MIA PaCa-2)
Biochemical Genetics (2019)
-
Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival
Journal of Applied Genetics (2016)
-
Norcantharidin inhibits Wnt signal pathway via promoter demethylation of WIF-1 in human non-small cell lung cancer
Medical Oncology (2015)