Abstract
Leiomyoma and leiomyosarcoma share morphological features and smooth muscle differentiation, and both arise most frequently within the uterine corpus of middle-aged women. However, they are considered biologically unrelated tumors due to their disparate clinical, cytogenetic, and molecular features. MED12, the mediator complex subunit 12 gene, has been recently implicated as an oncogene in as many as 70% of sporadic uterine leiomyoma. In the present study, we show MED12 hotspot exon 2 mutations in extrauterine leiomyoma (3 of 19 cases) and in leiomyosarcoma (3 of 13 uterine cases). We also show that MED12 mutations are found in both primary and metastatic leiomyosarcoma. Immunoblotting studies demonstrated MED12 protein expression in 100% of leiomyomas (13) and leiomyosarcomas (20), irrespective of MED12 exon 2 mutation status or histological grade. These findings indicate that MED12 has oncogenic roles in a broad range of smooth muscle neoplasia, including tumors arising in extrauterine locations.
Similar content being viewed by others
Main
Seventy percent of women in the general population develop uterine leiomyomas,1 and ∼25% of women experience substantial symptoms from these tumors, leading to over 200 000 hysterectomies annually in the United States.1 By contrast, uterine leiomyosarcomas are infrequent, with an estimated incidence of 0.64 per 100 000 women. Nonetheless, uterine leiomyosarcomas are among the most common sarcomas, accounting for 2–5% of tumors of the uterine body.2, 3, 4 There is substantial morphological overlap between leiomyoma and low-grade spindle cell leiomyosarcoma, and diagnostic distinction between these entities relies on histopathological criteria, including nuclear atypia, mitotic activity, and tumor cell necrosis.5 Morphological variants and unusual features can further complicate the diagnosis between leiomyoma versus leiomyosarcoma, as in smooth muscle tumors of uncertain malignant potential that cannot be unequivocally classified as benign or malignant.6
Recently, somatic mutations affecting MED12 (the mediator complex subunit 12 gene) were discovered in ∼70% of sporadic uterine leiomyoma.7, 8 Mediator is a modular protein complex of 25 subunits that regulates RNA polymerase II-mediated transcription, thereby orchestrating cell development and survival in cooperation with CDK8.9, 10 MED12 is located on chromosome sub-band Xq13.1 and is composed of 45 exons, although all MED12 mutations thus far described in uterine leiomyoma have affected exon 2 exclusively, with mutation hotspots in codons 36–44.
In the present study, we analyzed leiomyomas and leiomyosarcomas of various biological behaviors and anatomical locations to better determine the relevance of MED12 exon 2 mutations across a broad spectrum of smooth muscle neoplasia. In these studies, we also determined whether MED12 expression is dysregulated in smooth muscle neoplasia compared with a normal myometrium control. These studies demonstrate that MED12 mutations are the first known oncogenic mechanism shared by uterine and extrauterine leiomyoma and uterine leiomyosarcoma.
Materials and methods
Tumor Samples
Thirty-two leiomyomas and 27 leiomyosarcomas were analyzed from 51 patients, including 37 females and 14 males (Table 1). The leiomyomas were uterine (13) or extrauterine (soft tissue=4; esophagus=3; pelvis=2; stomach=2; retroperitoneum=2; and ovary, lung, kidney, bladder, urethra, and epididymis=1 each). The leiomyosarcomas included primaries of uterine (4)4 and extrauterine (6)6 locations and metastases from uterus (11)11 or extrauterine primaries (7).7 Histopathological and immunophenotypical features were reviewed by two experienced surgical pathologists. The tumor arising within the kidney was diagnosed as leiomyoma based on morphological features, negative HMB45 and S100 immunohistochemistry, and strong desmin expression. Immortal cell lines were established from three leiomyosarcoma biopsies: LMS03 was from a metastatic diaphragmatic nodule in a patient with primary thigh leiomyosarcoma; LMS04 was from a metastatic retroperitoneal lesion in a patient with primary uterine leiomyosarcoma; and LMS05 was from a primary thigh leiomyosarcoma. The SK-LMS-1 leiomyosarcoma cell line, from a primary vulvar leiomyosarcoma, was obtained from ATCC. The study was approved by the Institutional Review Board at Brigham and Women’s Hospital.
Sequencing
DNA was extracted from cell lines and fresh frozen tissue samples using a QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. PCR was performed with PCR SuperMix (Invitrogen, 11306), as follows: 1 cycle at 94 °C for 2 min followed by 35 cycles of 94 °C for 0.5 min, 60 °C for 0.5 min, 72 °C for 0.5 min, and a final extension at 72 °C for 5 min. Primers flanking MED12 exon 2 were from Makinen et al.:7 F: GCCCTTTCACCTTGTTCCTT, R: TGTCCCTATAAGTCTTCCCAACC. PCR products were evaluated by ethidium bromide staining on a 1% agarose gel alongside 1 Kb Plus DNA Ladder (Invitrogen). The PCR products were purified using QIAquick PCR Purification Kit (Qiagen), and Sanger sequenced. NM_005120.2 from NCBI was used as reference sequence.
Western Blotting Analysis
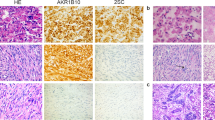
MED12 protein expression was analyzed in 37 tumors: 11 uterine leiomyomas, 2 extrauterine leiomyomas, 11 uterine leiomyosarcomas (4 primary, 7 metastatic), and 8 extrauterine leiomyosarcomas (3 primary and 5 metastatic). Frozen tumor samples were diced in ice-cold lysis buffer containing protease inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride), homogenized using a Tissue Tearor (Biospec Products, USA), and immunoblotted. Total protein lysate from non-neoplastic myometrium was positive control for MED12 expression; total protein lysate from GIST882 cells was used as exposure control. Uniform protein transfer was demonstrated by Ponceau S staining (Sigma Chemical), and immunostains were performed for MED12 (A300-774A, Bethyl Labs), mTOR (2972, Cell Signaling)—serving as a control at molecular mass similar to MED12—and GAPDH (G8795, Sigma). Detection was by chemiluminescence (Immobilon Western, Millipore), and signals were captured and quantified using a FUJI LAS1000plus system with Science Lab 2001 MultiGauge 2.3 software (Fujifilm Medical Systems, Stamford, CT, USA).
Results
MED12 mutations were demonstrated in 12 of 32 leiomyomas (37%), of which 9 were uterine and 3 were extrauterine (1 each from ovary, kidney, and retroperitoneum) (Table 1, Figures 1 and 2). In addition, MED12 mutations were demonstrated in 3 of 27 leiomyosarcomas (11%), all of uterine origin, one being a uterine primary, whereas the others were metastases from uterine primaries (Figures 1 and 2). One of the MED12-mutant leiomyosarcomas was analyzed as successive metastases, both of which contained the MED12 mutation (cases 41 and 42). None of the four leiomyosarcoma cell lines harbored MED12 mutations. None of the 10 extrauterine leiomyosarcomas had MED12 exon 2 mutations.
(a) MED12 exon 2 electrophorograms for six MED12-mutant leiomyomas (left) and leiomyosarcomas (right). (b) Schematic representation of the MED12 protein, highlighting distribution of mutations described to date in uterine leiomyoma, prostate cancer, and germline syndromes. MED12 mutations reported in this study are shown at bottom. MED12 protein domains are L: leucine-rich domain; LS: leucine- and serine-rich domain; PQL: proline-, glutamine-, and leucine-rich domain; OPA: opposite paired domain.
The 15 MED12 mutations affected either codon 44 (c.130-132) or, less frequently, codon 36 (c.106-108; Table 1). Ten tumors had missense point mutations, five had in-frame deletions, and one had a 39-nucleotide duplication at the intron 1–exon 2 boundary, predicted to create a 13 amino-acid insertion after codon 34 (Table 1). All mutations preserved the MED12 open reading frame.
MED12 protein was expressed in all leiomyomas and leiomyosarcomas, irrespective of whether they were mutant or wild type for MED12 exon 2. Leiomyoma and leiomyosarcoma MED12 expression levels ranged from 0.3- to 2.0-fold and from 0.4- to 2.7-fold, respectively, of those in normal myometrium (Supplementary Figure S1). The variations in MED12 expression levels within these narrow ranges did not correlate with MED12 mutation status, tumor location (uterine versus extrauterine), or tumor type (leiomyoma versus leiomyosarcoma).
Discussion
Initial reports highlighted the remarkable frequency of MED12 exon 2 mutations in uterine leiomyoma;7, 8, 11 and similar mutations were reported recently in 2 of 15 uterine leiomyosarcomas but not in 38 extrauterine leiomyosarcomas.12, 13 Likewise, we find MED12 mutations in 3 of 15 uterine leiomyosarcomas, including two tumors metastatic from uterine primaries, but not in 14 extrauterine leiomyosarcomas. All the mutant cases in our series affected females, consistent with a higher frequency of MED12 exon 2 mutations in uterine tumors. In all, we evaluated the MED12 exon 2 hotspot mutation region in 59 smooth muscle tumors, inclusive of both uterine and extrauterine leiomyomas and leiomyosarcomas. Notably, we identify MED12 mutations—for the first time—in extrauterine leiomyomas, where mutations were found in 3 of 19 cases (one each from ovary, kidney, and retroperitoneum). These advances show that MED12 oncogenic mutation is relevant across a broad spectrum of smooth muscle neoplasia and confirm that MED12 dysregulation provides an opportunity for evaluating oncogenic mechanisms shared by leiomyoma and leiomyosarcoma.
MED12 biological roles have not been characterized in leiomyoma and leiomyosarcoma, but none of the MED12 exon 2 mutations reported to date are destructive of the open reading frame, and many are missense mutations clustering to codons 36 and 44. This mutation profile is typical of an oncogene and indeed would be unprecedented in a classical tumor suppressor—wherein one would expect nonsense and frame-shift mutations, and less clustering to hotspots. Therefore, recent (non-quantitative) PCR and immunohistochemical evidence, showing downregulation of MED12 expression in leiomyosarcoma, is surprising.13 However, using quantitative assessment of MED12 proteins by immunoblotting, we demonstrate that both native and mutant MED12 expression levels are comparable, in leiomyoma and leiomyosarcoma, with normal MED12 expression levels in myometrium (Figure 3). Distinction between oncogenic and tumor suppressor roles could be relevant therapeutically, given that MED12 is necessary for CDK8 activation,14 and therefore MED12 gain-of-function oncogenic mutations could potentially be countered by CDK8 inhibitors. Despite the prevalence of leiomyoma, there are few effective medical therapies and hysterectomy remains a frequent treatment strategy, with considerable morbidities.15 Likewise, there exist no highly effective targeted therapies for leiomyosarcoma, which is an aggressive and often lethal sarcoma.
Despite the histological and immunophenotypical commonalities between leiomyoma and leiomyosarcoma, it is striking that the tumorigenic mutation mechanisms reported previously in leiomyoma are neither found generally in leiomyosarcoma nor are typical leiomyosarcoma mutations found in leiomyoma (Figure 4). Cytogenetic and molecular studies show distinct non-overlapping genetic pathways for leiomyoma and leiomyosarcoma,16 and malignant progression from leiomyoma to leiomyosarcoma is exceedingly rare, suggesting that leiomyoma is not on a biological continuum, oncogenically, with leiomyosarcoma. Leiomyomas typically have normal or noncomplex karyotypes in the diploid range, with highly recurrent clonal aberrations that include 7q deletion and HMGA2-region rearrangement.1 By contrast, most leiomyosarcomas are aneuploid with complex numerical and structural chromosomal aberrations and with substantial cytogenetic heterogeneity within and between cases. Molecular genetic studies also demonstrate divergent tumorigenic mechanisms in leiomyoma versus leiomyosarcoma (Figure 4). Recurrent oncogenic events in leiomyoma, beyond the above-mentioned rearrangements of high-mobility group genes HMGA2 and HMGA1, include fumarate hydratase gene (FH) mutations in syndromic leiomyomatosis17 and COL4A6 deletion in a variant of Alport syndrome with esophageal and vulvar leiomyomatosis (OMIM 308940). None of these genes has convincing tumorigenic roles in leiomyosarcoma, and the tumor genes implicated thus far in leiomyosarcoma, including TP53, MDM2, CDKN2A, RB1, and JUN, are ubiquitous oncogenes and tumor suppressors in human cancers rather than being specific for smooth muscle neoplasia.18
Nonetheless, occasional cases of leiomyosarcoma are reported to arise from leiomyoma,19, 20, 21, 22 and the possibility of biological continuum between certain leiomyoma and leiomyosarcoma is suggested by existence of histologically and clinically intermediate lesions—including cellular leiomyoma, atypical leiomyoma, leiomyoma with increased mitotic activity, STUMP, intravascular leiomyomatosis, and benign metastasizing leiomyoma. These observations suggest that certain uncommon leiomyoma subtypes have potential for progression to bona-fide leiomyosarcoma. The higher-risk leiomyoma subtypes include those with variant histologies and with certain genomic alterations such as translocation t(10;17) or 1p deletion.23, 24 Against this somewhat controversial backdrop, the present MED12 findings provide the first compelling oncogenic link between leiomyoma and leiomyosarcoma, even if this evidence does not prove that leiomyoma can progress to leiomyosarcoma.
In addition to the emerging role of exon 2 mutations in smooth muscle tumors, other MED12 mutations have been implicated in human disease. Germline mutations affecting MED12 exons 21 and 22—which encode a leucine- and serine-rich domain—cause the X-linked recessive hereditary syndromes Opitz–Kaveggia (OMIM 305450) and Lujan–Fryns (OMIM 309520).25, 26, 27 These disorders are characterized by overlapping phenotypes, including mental retardation and dysmorphic features, but are not associated with tumor predisposition;28 however, missense mutations of the same MED12 leucine- and serine-rich domain are found in 5% of prostate adenocarcinomas.29 The different MED12 mutation hotspots in smooth muscle neoplasia versus prostate cancer are consistent with multifaceted MED12 roles in various cell lineages and with the complex biology of the Mediator complex. Nonetheless, the possibility of MED12 mutations outside of exon 2 have not been evaluated to date, and additional studies are warranted to assess MED12 genomic and functional integrity, more comprehensively, in leiomyosarcoma.
In summary, MED12 exon 2 mutations are frequent oncogenic mechanisms in uterine leiomyoma and, albeit less frequently, in extrauterine leiomyoma and uterine leiomyosarcoma. MED12 mutation is the first recurrent oncogenic mechanism demonstrated in both benign and malignant smooth muscle tumors, and hence appears to have general relevance in neoplasms with smooth muscle differentiation, irrespective of histological grade. Further studies should enable characterization of the biological roles of MED12 oncoprotein and determine whether MED12-mediated oncogenic consequences can be inhibited therapeutically.
References
Walker CL, Stewart EA . Uterine fibroids: The elephant in the room. Science 2005;308:1589–1592.
Lin JF, Slomovitz BM . Uterine sarcoma 2008. Curr Oncol Rep 2008;10:512–518.
Amant F, Coosemans A, Debiec-Rychter M et al. Clinical management of uterine sarcomas. Lancet Oncol 2009;10:1188–1198.
Zaloudek C, Hendrickson MR . Mesenchymal tumors of the uterus In: Kurman RJ eds. Blaustein's Pathology of the Female Reproductive Tract. Springer-Verlag: New York, 2002, pp 561–615.
Bell SW, Kempson RL, Hendrickson MR . Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases Am J Surg Pathol 1994;18:535–558.
Schürch W, Bégin LR, Seemayer TA et al. Pleomorphic soft tissue myogenic sarcomas of adulthood. A reappraisal in the mid-1990s. Am J Surg Pathol 1996;20:131–147.
Makinen N, Mehine M, Tolvanen J et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011;334:252–255.
McGuire MM, Yatsenko A, Hoffner L et al. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS One 2012;7:e33251.
Bourbon HM, Aguilera A, Ansari AZ et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 2004;14:553–557.
Westerling T, Kuuluvainen E, Mäkelä TP . Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 2007;27:6177–6182.
Markowski DN, Bartnitzke S, Loning T et al. MED12 mutations in uterine fibroids-their relationship to cytogenetic subgroups. Int J Cancer 2012;131:1528–1536.
Je EM, Kim MR, Min KO et al. Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer 2012;131:E1044–E1047.
Perot G, Croce S, Ribeiro A et al. MED12 alterations in both human benign and malignant uterine soft tissue tumors. PLoS One 2012;7:e40015.
Knuesel MT, Meyer KD, Donner AJ et al. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol 2009;29:650–661.
Stewart EA . Uterine fibroids and evidence-based medicine – not an oxymoron. N Engl J Med 2012;366:471–473.
Quade BJ, Wang TY, Sornberger K et al. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer 2004;40:97–108.
Lehtonen R, Kiuru M, Vanharanta S et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol 2004;164:17–22.
Yang J, Du X, Chen K et al. Genetic aberrations in soft tissue leiomyosarcoma. Cancer Lett 2009;275:1–8.
Mittal K, Popiolek D, Demopoulos RI . Uterine myxoid leiomyosarcoma within a leiomyoma. Hum Pathol 2000;31:398–400.
Mittal KR, Chen F, Wei JJ et al. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod Pathol 2009;22:1303–1311.
Bukar M, Audu BM, Mustapha Z et al. Uterine leiomyosarcoma arising from a fibroid. J Obstet Gynaecol 2009;29:169–170.
Yanai H, Wani Y, Notohara K et al. Uterine leiomyosarcoma arising in leiomyoma: clinicopathological study of four cases and literature review. Pathol Int 2010;60:506–509.
Mittal K, Joutovsky A . Areas with benign morphologic and immunohistochemical features are associated with some uterine leiomyosarcomas. Gynecol Oncol 2007;104:362–365.
Hodge JC, Morton CC . Genetic heterogeneity among uterine leiomyomata: insights into malignant progression. Hum Mol Genet 2007; 16 Spec No, 1:R7–13.
Schwartz CE, Tarpey PS, Lubs HA et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet 2007;44.
Rump P, Niessen RC, Verbruggen KT et al. A novel mutation in MED12 causes FG syndrome (opitz-kaveggia syndrome). Clin Genet 2011;79:183–188.
Risheg H, Graham JM, Clark RD et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet 2007;39:451–453.
Lyons MJ . MED12-related disorders In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP (eds) GeneReviews. University of Washington: Seattle: Seattle (WA), 1993.
Barbieri CE, Baca SC, Lawrence MS et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685–689.
Acknowledgements
This study was supported by funding from the Daniel K. Ludwig Trust for Cancer Research (GR, AM-E, JS, GE, YW, MZ, and JF); a Sarcoma Alliance for Research Through Collaboration (SARC) Career Development Award (AM-E); and by the Department of Pharmacology, Universitá di Bologna, Italian Society of Pharmacology, and Fondazione del Monte di Bologna e Ravenna (all to GR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Ravegnini, G., Mariño-Enriquez, A., Slater, J. et al. MED12 mutations in leiomyosarcoma and extrauterine leiomyoma. Mod Pathol 26, 743–749 (2013). https://doi.org/10.1038/modpathol.2012.203
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.203
Keywords
This article is cited by
-
Recurrent broad ligament leiomyosarcoma with pancreatic and thigh metastasis: a case report
BMC Surgery (2020)
-
Myome – was liegt da in der DNA?
Der Gynäkologe (2019)
-
MED12, TERT promoter and RBM15 mutations in primary and recurrent phyllodes tumours
British Journal of Cancer (2018)
-
Leiomyoma with bizarre nuclei: a morphological, immunohistochemical and molecular analysis of 31 cases
Modern Pathology (2017)