Abstract
Prostate cancer is the second cause of cancer-related death in men of the Western world. The potential prognostic role of the combined alterations in EGFR and PTEN in prostate cancer is not well established. It was the aim of the study to investigate this role. Prevalence of EGFR and PTEN somatic mutations, EGFR amplification and EGFR protein expression were investigated in a series of prostate adenocarcinomas, classified according to the current Gleason grading system. Mutational analysis revealed eight EGFR and three PTEN mutations in 98 (8%) and 92 (3%) prostate adenocarcinomas, respectively. The combined prevalence of EGFR–PTEN mutations was 11%. EGFR overexpression was present in 31% of adenocarcinomas, with a marginally significant difference (P=0.068) between Gleason grade ≤7 adenocarcinomas and Gleason grade ≥8 and metastatic adenocarcinomas. Four cases (4 of 31; 13%) had an EGFR gene gain due to chromosome 7 polysomy. In 35% of adenocarcinomas we found some type of EGFR–PTEN alteration, with a tendency to be associated with advanced-stage prostate adenocarcinomas (P=0.04). The IVS18+19 polymorphism was also associated with more advanced prostate adenocarcinomas. This is the first study reporting mutations of EGFR and PTEN in the same series of prostate adenocarcinomas. Protein overexpression is the most frequent EGFR abnormality. Mutations in EGFR and PTEN genes are a minor event, although prostate cancer represents the third neoplasm in which the EGFR gene mutations are more prevalent. Alterations in the EGFR–PTEN signaling pathway are present in a third of prostate adenocarcinomas, particularly affecting the more advanced cases.
Similar content being viewed by others
Main
EGFR belongs to the ErbB family of tyrosine kinase receptors.1 They are activated by binding to peptide growth factors of the EGF family, leading to activation of multiple downstream pathways. The Ras-Raf-MEK-ERK1/2, STAT-3, and STAT-5, and the PI3K/PTEN-Akt-Mtor cascades are the main pathways activated downstream of ErbB receptors, which have strong regulatory effects on cell proliferation, differentiation, survival and migration.2, 3
In many different cancer cell types, the ErbB pathway becomes hyperactivated by a range of mechanisms.2, 4 Increased levels of EGFR gene expression have been reported in adenocarcinomas of the head and neck, lung, pancreas, bladder, ovary, cervix, brain, breast, colon and prostate, among others, and have been frequently associated to adverse prognosis.1, 3, 5, 6
EGFR gene amplification, as well as structural genetic alterations, has been reported in several adenocarcinoma types, including non-small-cell lung cancer (NSCLC), glioblastoma, pancreatic cancer, and head and neck squamous cell carcinoma. Point mutations or short DNA deletions have mainly been reported in lung cancer, with a low frequency in prostate and colorectal cancers and a very uncommon occurrence in head and neck, pancreas, esophagus and breast cancer, among others.3, 6, 7, 8, 9, 10, 11, 12
In prostate cancer, EGFR overexpression has also been described. Some studies report an increasing expression along the natural history of prostate cancer, as well as an association with poor prognostic factors such as high-grade, advanced-stage and progression to hormone refractory status.5, 13 Recently, EGFR point mutations and amplification have been found in prostate cancer, particularly in advanced adenocarcinomas.14, 15 However, the number of cases in these series is small, and on the other hand another recent study performing analysis of EGFR somatic mutations, amplification and protein expression in the same subsets of prostate adenocarcinomas did not find mutations.13
The PI3K–PTEN–AKT signaling pathway has been shown to be important in the survival and proliferation of human prostate cancer cells and to regulate prostate adenocarcinoma angiogenesis.16, 17 In response to growth factors, the PI3K pathway can be activated by different protein tyrosine kinases, including the ErbB family and FGFRs among others. The PTEN phosphatase, in turn, has an important role in the regulation of phosphatidylinositol-3-kinase (PI3K) product (PIP3), as a central negative regulatory factor of PI3K–AKT pathway.18, 19 PI3K–AKT pathway is one of the major signal transduction pathways that promote cell growth, survival and proliferation. In addition, PTEN is involved in cell-cycle progression and regulation of DNA damage key checkpoint proteins. Moreover, PTEN regulates p53 stability, and p53 in its turn positively controls PTEN transcription.20
PTEN alterations have been described in different human tumor types such as glioblastoma, breast cancer, lung cancer, melanoma and prostate cancer, among others. Somatic alterations of PTEN include point mutations, small deletions, homozygous deletions and epigenetic silencing.19, 21 Functional loss of PTEN and subsequent activation of the AKT pathway is one of the most frequent abnormalities in prostate cancer progression, and chromosomal region mapping has shown PTEN to be the most frequently deleted gene in prostate cancer.22 Different studies have indicated that PTEN haploinsufficiency could be an early prognostic marker for prostate cancer, and complete loss of PTEN expression appears to correlate with advanced pathologic stage and high Gleason score.21, 23, 24, 25 Somatic PTEN deletions and mutations have been described in primary and, more frequently, in metastatic prostate adenocarcinomas. The reported frequency of PTEN mutations in metastatic adenocarcinomas varies between 20 and 60%. In primary adenocarcinomas, lower frequencies have been reported, with discrepancy among the different studies, and an overall estimation of approximately 15%.26, 27, 28, 29, 30, 31 However, both the number of papers analyzing somatic mutational status of PTEN and their sample size are too small to establish their actual prevalence in primary prostatic adenocarcinomas.
The potential prognostic role of the alterations in EGFR and its downstream effector PTEN in prostate cancer is not well established. Several clinical strategies are available to block EGFR receptor-mediated oncogenic function. It would be very important to determine the different types of aberrant events in EGFR and PTEN, to understand the pathogenesis and natural history of prostate cancer and to select appropriate treatment strategies with different inhibitors. In this study, we have investigated the prevalence of EGFR and PTEN somatic genetic alterations and protein expression in a large group of well-defined prostate adenocarcinomas from a Caucasian population, classified by stage and Gleason score.
Materials and methods
Patients and Adenocarcinoma Samples
A total of 98 prostate adenocarcinomas were selected retrospectively from the files of the Department of Pathology, at the Hospital del Mar in Barcelona, Spain, and were the subject of this report. From these, 12 were needle biopsy, 61 radical prostatectomy adenocarcinomas, 16 adenocarcinomas found at autopsy, so-called latent adenocarcinomas, 3 found at cystoprostatectomy (incidental) and 6 bone metastases. All adenocarcinomas were staged with the TNM classification. According to the current Gleason grading criteria, the prostate tumors were classified as combined Gleason scores 6 (n=32), 7 (n=39) and ≥8 (n=21). 89 samples were formalin-fixed paraffin-embedded adenocarcinomas, and 9 samples were frozen adenocarcinomas. Although no information on grade was available from the respective primary adenocarcinomas, metastatic samples (n=6) were grouped together with Gleason score ≥8 tumors, assuming they belonged to an aggressive category.
The autopsy samples dated from 1993 to 2007. The remaining cases were recruited between 2000 and 2008.
EGFR and PTEN Mutation Analysis
DNA extraction and DNA quality checking was performed in all samples as previously described.32 Exons 18–21 of EGFR and exons 5–8 of PTEN were amplified and sequenced in 98 cases for EGFR and in 92 cases for PTEN using tumor DNA. We used exon-specific primers binding to adjacent intron sequences except for exons 5 and 8 of PTEN, in which the primer comprised a part of the exon analyzed. The primers for EGFR were: CCCTTGTCTCTGTGTTCTTG (forward) and TCATGGTCTGGTGGGGA (reverse) for exon 18; AGCATGTGGCACCATCTCAC (forward) and TCAGGCCCACCTTTTCTC (reverse) for exon 19; AGCCACACTGACGTGCCTCT (forward) and ATAAGGAGCCAGGATCCTCA (reverse) for exon 20 and TGCAGAGCTTCTTCCCATGA (forward) and AGGTGGCTTTAGGTCAGCCA (reverse) for exon 21. The primers for PTEN were CTTATTCTGAGGTTATCT (forward) and GGTGTAATGATATGTGCA (reverse) for exon 5A; CACTGTAAAGCTGGAAAGGGACG (forward) and TCCTCTTCCTGGATCTGAG (reverse) for exon 5B; TTTTCTGTCCACCAG (forward) and CCATGTATTGGAACAGT (reverse) for exon 6; CCTGTGAAATAATACTGGT (forward) and CATTGGGAGAAATATCCAA (reverse) for exon 7 and GTGATCAAGAAATCGATA (forward) and GGTTGTTGACTTGTATGT (reverse) for exon 8.
PCR reactions, product purification and mutation analysis were performed as previously described.32 PCR conditions were 95°C (5 min) for 1 cycle, 95°C (40 s), 63°C (40 s) for EGFR and 54°C (40 s) for PTEN, 72°C (40 s) for 40 cycles and a final extension step of 72°C (5 min). PCR products were separated by electrophoresis and visualized with ethidium bromide. Samples without DNA templates were included in all assays as negative controls. Each PCR product was sequenced in both forward and reverse directions. All mutations reported were confirmed with two independent rounds of PCR and direct sequencing. As quality control, independent PCR products were amplified and sequenced in 14% of wild-type (wt) cases to confirm the mutational results using DNA from the same adenocarcinoma areas.
Immunohistochemistry of EGFR and PTEN
Immunohistochemical staining for EGFR was performed on 75 cases, all of them from formalin-fixed paraffin-embedded tissue sections, using a highly specific mouse monoclonal antibody (Clone H11; Dako, Glostrup, Denmark). In all samples, immunoreaction was revealed using the EnVision Plus system (Dako) and counterstained with hematoxylin. EGFR expression was evaluated semiquantitatively by estimating the percentage of positive adenocarcinoma cells as follows: score 0, no detection of positive cells; 1+, weak positivity in <10% cancer cells; 2+, moderate or strong positivity in 10–40% cancer cells and 3+, moderate or strong positivity in >50% cancer cells. An adenocarcinoma was considered to overexpress EGFR when the score was 2+ or 3+.
Fluorescence in situ Hybridization Analysis of EGFR
To assess the genetic status of EGFR, we carried out fluorescence in situ hybridization (FISH) on formalin-fixed paraffin-embedded tissues, in the 31 cases in which there was tumor tissue available for this purpose. The EGFR probe consists of two probes, one with the centromeric α-satellite probe, specific for chromosome 7 (spectrum green), and a locus-specific probe for the EGFR gene, located at 7p12 (spectrum orange) (Abbott Molecular Inc., Des Plaines IL, USA). We serially performed a hematoxylin–eosin stain and FISH in consecutive tissue sections. After pathological confirmation, FISH slides were deparaffinized, pretreated with EDTA solution in a microwave and digested with pepsin (0.05 mg/ml in 0.01 N HCL) for 30 min at 37°C. Tissue morphology was assessed by phase-contrast microscopy to ensure sufficient digestion of the collagen matrix. Pretreated tissue sections and probes were co-denatured at 78°C for 5 min and hybridized overnight at 37°C in a hot plate (Hybrite chamber; Abbot Molecular Inc.). After hybridization washes, slides were counterstained with 4,6-diamino-2-phenylindole (DAPI counterstain; Abbott Molecular Inc.).
Results were analyzed in a fluorescence microscope (Olympus; BX51) using the CytoVision software (Applied Imaging, Santa Clara, CA, USA). Tissue sections were scanned at low magnification ( × 100) with DAPI excitation. A minimum of 100 nuclei were scored by two observers. Based on previous publications,13 an adenocarcinoma was considered amplified if the EGFR/centromere 7 ratio was ≥2.0. We used adjacent normal tissues from four cases as controls. The cutoff for polysomy (3—4 copies) was 7%.
Results
EGFR and PTEN Mutational Analyses
EGFR mutations were sought in 98 and PTEN mutations in 92 prostate adenocarcinoma samples. All the cases analyzed for PTEN were also tested for EGFR. Of 98, 8 (8.2%) adenocarcinomas harbored EGFR mutations, and of 92 adenocarcinomas, 3 (3.3%) had PTEN mutations (Table 1).
All EGFR mutations were located in exons 20 and 21. Two of them were found in Gleason score 6 adenocarcinomas (2 of 32; 6.3%) and 6 of them in Gleason ≥7 adenocarcinomas (6 of 66; 9.1%). We identified 5 mutations: I821T and I789I in exon 20, and L858L, H870N and V834A in exon 21. The most common mutation was V834A substitution, present in three adenocarcinomas. Only the L858L silent change had been previously reported (Figure 1).
EGFR and PTEN sequence analysis in prostate tumors. Case 1 presented a mutation in exon 20, transition T → C at codon 821. Case 2 presented a mutation in exon 21, transition A → G at codon 834. Case 3 presented a deletion of an adenine at codon 164 in exon 5. Case 4 presented a mutation in exon 7, transition C → T at codon 223.
We also found two polymorphisms: Q787Q in exon 20 and IVS18+19 in intron 18. Both of them have been described previously in normal population (http://www.ncbi.nlm.nih.gov/SNP) with frequencies of 43 and 3%, respectively, in heterozygosity. The Q787Q polymorphism was detected in 69 cases (70.4%), and the intron 18 polymorphism IVS18+19 was detected in 17 cases (17.3%). Fifteen prostate adenocarcinomas presented both polymorphisms in the same sample. Thus, the prevalence of both polymorphisms was higher in our series of prostate adenocarcinomas than in the SNPs database; 17.3 vs 3% for IVS18+19 and 70.4 vs 43% for Q787Q. Distribution of adenocarcinoma polymorphisms according to the Gleason score classification is shown in Figure 2. The presence of IVS18+19 was statistically associated with more aggressive prostate adenocarcinomas (Gleason ≥8 adenocarcinomas and metastases) (Fisher's exact test P=0.042). With exception of two cases, all adenocarcinomas with IVS18+19 presented also Q787Q polymorphism.
Only 3 of 92 adenocarcinomas (3.3%) showed PTEN mutations. Two of them carried missense mutations, P95S (exon 5) and D223N (exon 7), and one had a deletion of an adenine leading to a premature stop in codon 164 (exon 5) (Figure 1). Both cases with missense mutations were Gleason score 7 adenocarcinomas, and the case with the PTEN truncating mutation was a Gleason score 10 adenocarcinoma.
Taking together mutational analyses of both genes, 11 (11.6%) adenocarcinomas presented a genetic alteration in the EGFR–PTEN signaling pathway. Presence of PTEN and EGFR somatic alterations seemed to be mutually exclusive, because none of the adenocarcinomas harbored mutations in both genes.
For the statistical analysis, we have excluded three cases in which it was not possible to perform PTEN mutational study and EGFR was wt. If we analyze the overall prevalence of EGFR and PTEN mutations according to Gleason score, 2 of 32 (6.3%, Gleason score 6), 6 of 38 (16%, Gleason score 7) and 3 of 25 (12%, Gleason score ≥8 and metastases) had a mutation. Prevalence of mutations in Gleason score 7 and >7 adenocarcinomas was similar, so considering only two groups (adenocarcinomas with Gleason score 6 vs adenocarcinomas with Gleason score ≥7), 2 of 32 (6.3%) adenocarcinomas with Gleason score 6 presented a mutation vs 9 of 63 (14.3%) adenocarcinomas with Gleason score ≥7. Although, this difference is not statistically significant, it seems that mutations in EGFR–PTEN signaling pathways may be more frequent in high Gleason score adenocarcinomas.
EGFR and PTEN immunohistochemistry
In 75 adenocarcinomas we analyzed the protein expression using immunohistochemistry. Normal, atrophic and hyperplastic prostate glands showed EGFR protein expression in basal cells. Neoplastic prostate glands had no expression in most of the cases (n=48), whereas EGFR immunostaining was detected in 27 adenocarcinomas. EGFR overexpression was considered when adenocarcinoma cells showed an immunohistochemical score from 2+ to 3+. Thus, 23 adenocarcinomas (31%) overexpressed EGFR. In 4 of 27 adenocarcinomas we found a very weak expression, classified as score 1+, and we grouped them with the negative cases. Nine adenocarcinomas were considered as score 2+, and 14 cases showing very strong EGFR expression were classified as score 3+. EGFR-overexpressing adenocarcinomas belonged to different Gleason score groups: 6 of 28 (21.4%) were Gleason score 6, 8 of 28 (28.6%) Gleason score 7 and 9 of 19 (47.4%) Gleason score ≥8 and metastases (Figure 3). Comparing adenocarcinomas with Gleason score ≤7 and adenocarcinomas with Gleason score ≥8 and metastases, 14 of 56 (25%) vs 9 of 19 (47.4%) showed EGFR overexpression (Pearson's χ2-test, P=0.068), indicating that there is a statistical trend toward EGFR overexpression to be associated with the more aggressive prostate adenocarcinomas. In 75 samples, both mutation and protein expression analyses were performed. There was no statistical association between the presence of mutation and EGFR overexpression.
Examples of immunohistochemical expression of EGFR in prostate adenocarcinomas. (a) Gleason score 9, wild-type prostate adenocarcinoma showing moderate to strong EGFR expression. (b) Gleason score 9 bone metastatic adenocarcinoma, wild-type, showing moderate EGFR expression. (c) Absence of EGFR expression in a V834A bone metastatic mutant adenocarcinoma (original magnification, × 200).
FISH Analysis of EGFR
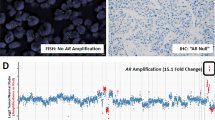
EGFR gene amplification was investigated by FISH analysis in 31 adenocarcinomas, 13 cases were Gleason score 6 and 14 were Gleason score ≥7 and 4 were Gleason score ≥8 and metastases. None of the 31 cases showed EGFR amplification, being EGFR/centromere 7 ratios <1.5. Four cases (4 of 31; 13.3%) had gain of EGFR gene due to chromosome 7 polysomy. All of these cases had 3–5 copies of chromosome 7 in 30–70% of the cells (Figure 4).
Although the number of cases with EGFR gain is low and we have not found statistical significance (Fisher's exact test, P=0.120), the presence of polysomy seems to be related with high-grade adenocarcinomas, as all cases with chromosome 7 gain were Gleason score ≥7.
In 23 adenocarcinomas, immunohistochemistry of both EGFR and FISH analyses could be performed. There was no relationship between EGFR protein expression and gene copy number tested using FISH analysis: the five EGFR-overexpressing adenocarcinomas in this subgroup did not show EGFR gain and one adenocarcinoma with polysomy did not show EGFR immunostaining. No information regarding immunohistochemical expression was available for the other three cases with EGFR gain due to polysomy. However, the number of cases studied for both analyses was too low to draw definitive conclusions.
Total EGFR and PTEN Alterations
Including as EGFR alterations somatic mutation, copy number gain and/or immunohistochemical expression, 31 adenocarcinomas presented an aberrant event in the EGFR molecule or gene (Figure 5). The most frequent aberrant activating event was EGFR protein overexpression, present in 31.1% of prostate adenocarcinomas analyzed by immunohistochemistry. In 75 cases we performed both mutation and immunohistochemistry analyses, and in 30 cases we performed both mutation and FISH analyses. Overexpression and mutation were coexistent in only four cases. Nevertheless, taking into account that we have combined information on the three analyses in 23 adenocarcinomas only, it has not been possible to determine whether the different types of EGFR alterations are coexistent or exclusive. In any event, we tested the distribution of total EGFR aberrant events in our series of prostate adenocarcinomas. If we classify the altered EGFR adenocarcinomas according to their Gleason score, 8 of 32 Gleason score 6 adenocarcinomas (25%), 11 of 38 Gleason score 7 adenocarcinomas (29%) and 12 of 28 Gleason score ≥8 and metastases (43%) presented an EGFR alteration.
Comparing adenocarcinomas with Gleason scores ≤7 and >7, we found that the prevalence of alterations was 27 vs 43%. Taking together total EGFR alterations and PTEN mutations, we found that 34 adenocarcinomas (35%) presented an aberrant event in the EGFR–PTEN signaling pathway. However, for the statistical analysis, we excluded negative cases in which it was not possible to test EGFR or PTEN alterations by all the techniques. Thus, in 52 adenocarcinomas we could test the association between immunohistochemistry and FISH positivity, mutations and Gleason classification. Alterations in the EGFR–PTEN signaling were related to an increasing Gleason score of the adenocarcinomas. Eight of 17 (47%) Gleason score 6, 13 of 20 (65%) Gleason score 7 and 13 of 15 (86.7%) of Gleason score ≥8 and metastases presented EGFR overexpression or EGFR copy number gain or somatic mutations (Pearson's χ2-test, P=0.063). Comparing Gleason score ≤7 adenocarcinomas with Gleason score ≥8 adenocarcinomas and metastases, we found that the presence of alterations in EGFR or PTEN was statistically associated with higher-grade prostate adenocarcinomas (Pearson's χ2-test, P=0.04; Figure 6).
For the statistical analysis, we excluded negative cases in which it was not possible to test EGFR or PTEN alterations by all the techniques. Comparing Gleason score ≤7 with Gleason score ≥8 and metastases, EGFR–PTEN alterations are statistically associated with higher-grade prostate adenocarcinomas (Pearson's χ2-test, P=0.04).
Relationship between EGFR Status and Biochemical Recurrence
Of the 49 patients with available PSA data and a follow-up of at least 24 months, biochemical recurrence was detected in 13, whereas in the remaining 36 cases there was no sign of biochemical or clinical recurrence at last follow-up. Fourteen cases in this subgroup with available PSA data showed some type of EGFR alteration and one presented a PTEN mutation, whereas the remaining 34 cases were wt. No statistical association between EGFR–PTEN mutations and recurrence, nor between EGFR overexpression and biochemical PSA values was found, although only 8% of cases without EGFR overexpression showed biochemical recurrence, whereas 30% of patients with EGFR-overexpressing adenocarcinomas did. In addition, there was no statistical association between the polymorphic variants and recurrence, although the percentage of patients who showed biochemical PSA increase at follow-up was higher in cases with the IVS18+19 variant (36.4%, corresponding to 4 of 11 cases with the polymorphism) compared with those without it (23.7%, or 9 from 29 cases without the polymorphism).
Finally, there was a significant association between combined Gleason score and biochemical recurrence, as about 67% of Gleason score ≥8 and metastatic patients developed biochemical recurrence, whereas only 24% of Gleason score 7 and none of Gleason score 6 patients did recur (Fisher's exact test P<0.0001). These results are an indication that this series of patients was not biased and reflected the normal spectrum of the disease.
Discussion
The molecular changes involved in the pathogenesis and the natural history of prostate adenocarcinoma are poorly understood. Thus, the essential steps that mark the transition from the early phases of prostate cancer development to more aggressive stages of the disease are not known. The main parameters guiding therapy and prognosis are still pathological grade and stage. There is a need for new prognostic and therapeutic markers, particularly with the development of new molecular-targeted therapies, such as the anti-EGFR molecules. In this study, we have investigated the changes in EGFR and PTEN in different clinical–pathological subsets of prostate cancer.
Increased levels of EGFR gene expression have been detected in a high variety of human adenocarcinomas including prostate cancer, mainly using immunohistochemical techniques, and they have been frequently related to adverse prognosis. However, whether EGFR overexpression is a prognostic factor in patients with these adenocarcinomas is still controversial.1, 3, 6 Till date, very few reports have analyzed the EGFR mutational status in prostate cancer. In fact, only three articles have reported the presence of EGFR somatic mutations in this type of adenocarcinoma, all of them based on a small number of cases. To date, the prevalence of EGFR somatic mutations in prostate cancer published in the Catalogue of Somatic Mutations in Cancer (www.sanger.ac.uk) is about 6%. Douglas et al15 identified the presence of EGFR mutations in prostate cancer for the first time, and they found four not previously reported missense mutations in the tyrosine kinase domain of EGFR. The prevalence of mutations in this study was 4.5% (4 of 89 patients). Interestingly, the samples analyzed belonged to Caucasian, African American and Korean patients, and three of the four mutated adenocarcinomas were from Korean patients. Later on, Cai et al33 reported that three of these four mutations were constitutively active oncogenic mutations. Schlomm et al13 also analyzed the EGFR exons 18–21, but did not find mutations, although the number of adenocarcinomas analyzed was very low. Cho et al14 reported nine EGFR mutations in three advanced prostate cancer cases, and they were not confirmed by additional independent rounds of PCR amplification and sequencing.
To the best of our knowledge, this is the first study in which the significance of the EGFR and PTEN alterations in prostate cancer is investigated focusing on the relationship among somatic mutations, gene amplification and protein expression. In addition, this is the largest series of prostate cancer cases in which EGFR mutations have been analyzed. Moreover, this is the first study reporting mutations of EGFR and PTEN in the same series of prostate adenocarcinomas. Our study data indicate a prevalence of 8.2% for EGFR and 3.3% for PTEN mutations. Taking together both genes, 11.3% prostate adenocarcinomas presented somatic point alterations in the EGFR–PTEN signaling pathway. The prevalence of mutations was higher in aggressive prostate adenocarcinomas (Gleason score ≥7), although this trend was not statistically significant. The presence of EGFR and PTEN mutations seemed to be mutually exclusive because none of the adenocarcinomas harbored mutations in both genes.
PTEN mutation prevalence in our series is lower than that reported previously. We have found only three PTEN mutations, two of them had been previously described in other tumor types34, 35 and the missense mutation D223N has not been reported before. In the prostate cancer literature, PTEN mutations have been described with discrepant data.26, 27, 28, 29, 30 It is noteworthy that although PTEN mutations were reported in prostate cancer for the first time in 199736 relatively few papers have analyzed the mutational status of this gene in prostate cancer since then, and perhaps the number of adenocarcinomas tested in these studies has been too small to establish the true prevalence of somatic mutations in primary prostate cancer.
In the EGFR mutational analysis, the percentage of mutations is also low, about 8%. Nevertheless, the percentage of EGFR mutations reported in the literature on human adenocarcinomas is also low. The highest incidence of EGFR mutations in Caucasian patients (10%) has been reported in NSCLC, followed by adrenocortical carcinoma (9%) (http://www.sanger.ac.uk/). According to our study results, prostate cancer would be the third most frequent neoplasm harboring somatic EGFR mutations.
Most of EGFR mutations described in human adenocarcinomas are located in the tyrosine kinase domain, affecting exons 18–21. The in-frame deletion in exon 19 involving codons 746–753, and a missense mutation leading to a substitution at codon 858 (L858R) in exon 21, are two mutation hotspots, which account for more than 90% of all detected mutations in lung cancer.37, 38, 39, 40 Codon 858 is the most frequently mutated hotspot for EGFR in human cancer, and missense mutation L858R is almost exclusive of lung cancer, where it constitutes around 42% of EGFR mutations (http://www.sanger.ac.uk/).
In our series of prostate adenocarcinomas we have found mutations in exons 20 and 21 of EGFR. Although the L858L change had already been reported in upper aerodigestive tract adenocarcinomas, the remaining mutations are described for the first time in this article. Interestingly, codons 834 and 870, which contained mutations in four prostatic adenocarcinomas in our series, have been found to be mutated in lung, thyroid and head and neck adenocarcinomas, but with different resulting amino-acid changes (http://www.sanger.ac.uk/). From previous reports in prostate cancer and other primary adenocarcinomas, data contained in the ‘Catalogue of Somatic Mutations in Cancer’ database (http://www.sanger.ac.uk/), and our own results, it appears that in lung cancer EGFR mutations occur mostly as hotspot somatic mutations, whereas in other adenocarcinoma types they are distributed in different codons along exons 18–21. The E709K missense mutation, found in two prostate adenocarcinomas by Cho et al14 could be considered an exception, as it has been found also in 11 lung cancer samples (http://www.sanger.ac.uk/).
Several authors have analyzed the immunohistochemical expression of EGFR in prostate cancer. There is a high variability of EGFR expression, ranging from 1 to 100% in different studies.5, 13, 41, 42 Data on the relationship between Gleason score and EGFR expression are also conflicting.43, 44, 45 In recent papers, there is more agreement in the association between EGFR immunostaining and advanced adenocarcinoma stage, high Gleason score and preoperative PSA. These results support a role for EGFR expression in the development of prostate cancer and in the progression to an androgen-independent, hormone-refractory clinical phase.5, 13, 42 Schlomm et al13 reported EGFR immunostaining in 18% of samples, in a large tissue microarray immunohistochemical study with more than 2000 adenocarcinomas. In addition, it has been suggested that EGFR overexpression is significantly less common in Caucasian (18%) than in African American (46%) and Korean (72%) patients.15, 41 We have found EGFR expression in 31% of adenocarcinomas, a higher percentage than that of previous reports based on Caucasian populations.13, 15, 41 Moreover, our study results concur with those of Schlomm et al13 in finding an association between increased EGFR expression and high Gleason score. They also performed mutational analysis in the same cases, with negative results.
Using FISH, Schlomm et al13 also found gains in EGFR copy number due to chromosome 7 polysomy in 3.3% of cases, whereas EGFR amplification was observed in 0.3% of cases. In addition, they found a significant association of EGFR copy number gains with protein expression, and with higher-grade and advanced-stage adenocarcinomas. In our series, EGFR gains were identified in 4 of 30 adenocarcinomas (13.3%), all of them with a low copy number increase (3–5 copies) due to chromosome 7 polysomy. All FISH-positive adenocarcinomas were Gleason score ≥7, and EGFR amplification was not found in any of them. Although there was no correlation between EGFR expression and EGFR copy number gains, a statistically significant association was found between increase in EGFR, due to either protein overexpression or chromosome 7 polysomy, and high Gleason score.
About 31% of prostate adenocarcinomas in the present series showed EGFR overactivation by mutation, overexpression and/or increased copy number. Taking together total EGFR alterations and PTEN mutations, 34 adenocarcinomas (35%) presented some sort of aberrant event in the EGFR–PTEN signaling pathway. Total alterations in EGFR–PTEN signaling were related to adenocarcinoma Gleason score, with a statistically significant association with Gleason score ≥8 and metastatic adenocarcinomas (Pearson's χ2-test, P=0.04).
PTEN mutation represents a minor aberrant event in the pathogenesis of prostate adenocarcinomas. It would be interesting to investigate other PTEN alteration mechanisms, such as deletion or epigenetic inactivation. The most common mechanism for PTEN inactivation of both alleles is mutation of one of them and deletion of the other, although reduced PTEN protein levels are often seen in the absence of genomic abnormalities.46, 47 Loss of heterozygosity (LOH) of PTEN has been frequently reported in primary prostate adenocarcinomas, but the proportion of adenocarcinomas with deletion of one allele and mutation of the other is low.29, 48 Dong et al26 reported, in a series of low-stage adenocarcinomas, that PTEN LOH and somatic mutations were uncommon, and they concluded that alteration of PTEN was not an early event in prostatic carcinogenesis. Conversely, different studies have indicated that PTEN haploinsufficiency could be an early prognostic marker for prostate cancer, and complete loss of expression appears to correlate with pathological markers of poor prognosis and tumor progression.20, 21, 23, 25, 30, 49 Mouse models for prostate cancer also suggest that PTEN may have a role in initiation or early progression of the disease.18, 37, 50, 51 Interestingly, adenocarcinomas from heterozygous PTEN(+/−) mice lose the wt PTEN allele, implying that adenocarcinoma formation requires lack of both PTEN alleles.24
To the best of our knowledge this is the first paper and the largest series analyzing EGFR and PTEN mutations in prostate adenocarcinomas from Caucasian population, and also the first to report EGFR and PTEN mutations in the same series of prostate adenocarcinomas. In addition, we have also tested protein expression by immunohistochemistry and EGFR copy number by FISH. We conclude that the EGFR signaling pathway is involved in about one third of prostate adenocarcinomas, particularly in high Gleason score adenocarcinomas. Protein overexpression is the most frequent type of EGFR aberrant event. Furthermore, overrepresentation of EGFR due to chromosome 7 polysomy or protein overexpression is statistically associated with more aggressive prostate adenocarcinomas. Mutations in EGFR and PTEN are a minor event, although they are involved in about 11% of prostate cancers, and thus prostate cancer ranks third among adenocarcinomas with EGFR mutations. The use of tyrosine kinase inhibitors could be beneficial in the treatment of prostate cancer, however it is imperative to investigate other downstream regulatory elements of EGFR signaling, to choose the more adequate targets for effective cancer treatment.
References
Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2–16.
Yarden Y, Sliwkowski MX . Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127–137.
Sibilia M, Kroismayr R, Lichtenberger BM, et al. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation 2007;75:770–787.
Sebastian S, Settleman J, Reshkin SJ, et al. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta 2006;1766:120–139.
Di Lorenzo G, Tortora G, D’Armiento FP, et al. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin Cancer Res 2002;8:3438–3444.
Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–181.
Janne PA, Johnson BE . Effect of epidermal growth factor receptor tyrosine kinase domain mutations on the outcome of patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res 2006;12:4416s–4420s.
Kwak EL, Jankowski J, Thayer SP, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res 2006;12:4283–4287.
Laurent-Puig P, Lievre A, Blons H . Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res 2009;15:1133–1139.
Lee J, Jang KT, Ki CS, et al. Impact of epidermal growth factor receptor (EGFR) kinase mutations, EGFR gene amplifications, and KRAS mutations on survival of pancreatic adenocarcinoma. Cancer 2007;109:1561–1569.
Lee JW, Soung YH, Kim SY, et al. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res 2005;11:2879–2882.
Nagahara H, Mimori K, Ohta M, et al. Somatic mutations of epidermal growth factor receptor in colorectal carcinoma. Clin Cancer Res 2005;11:1368–1371.
Schlomm T, Kirstein P, Iwers L, et al. Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin Cancer Res 2007;13:6579–6584.
Cho KS, Lee JS, Cho NH, et al. Gene amplification and mutation analysis of epidermal growth factor receptor in hormone refractory prostate cancer. Prostate 2008;68:803–808.
Douglas DA, Zhong H, Ro JY, et al. Novel mutations of epidermal growth factor receptor in localized prostate cancer. Front Biosci 2006;11:2518–2525.
Fang J, Ding M, Yang L, et al. PI3K/PTEN/AKT signaling regulates prostate adenocarcinoma angiogenesis. Cell Signal 2007;19:2487–2497.
Gao N, Zhang Z, Jiang BH, et al. Role of PI3K/AKT/mTOR signaling in the cell cycle progression of human prostate cancer. Biochem Biophys Res Commun 2003;310:1124–1132.
Di Cristofano A, Pandolfi PP . The multiple roles of PTEN in adenocarcinoma suppression. Cell 2000;100:387–390.
Jiang BH, Liu LZ . PI3K/PTEN signaling in adenocarcinomaigenesis and angiogenesis. Biochim Biophys Acta 2008;1784:150–158.
Liu W, Zhou Y, Reske SN, et al. PTEN mutation: many birds with one stone in adenocarcinomaigenesis. Anticancer Res 2008;28:3613–3619.
Chow LM, Baker SJ . PTEN function in normal and neoplastic growth. Cancer Lett 2006;241:184–196.
Liu W, Chang B, Sauvageot J, et al. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer 2006;45:1018–1032.
Kwabi-Addo B, Giri D, Schmidt K, et al. Haploinsufficiency of the Pten adenocarcinoma suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci USA 2001;98:11563–11568.
Schmitz M, Grignard G, Margue C, et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer 2007;120:1284–1292.
Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 2007;97:678–685.
Dong JT, Sipe TW, Hyytinen ER, et al. PTEN/MMAC1 is infrequently mutated in pT2 and pT3 carcinomas of the prostate. Oncogene 1998;17:1979–1982.
Dong JT . Prevalent mutations in prostate cancer. J Cell Biochem 2006;97:433–447.
Majumder PK, Sellers WR . Akt-regulated pathways in prostate cancer. Oncogene 2005;24:7465–7474.
Pesche S, Latil A, Muzeau F, et al. PTEN/MMAC1/TEP1 involvement in primary prostate cancers. Oncogene 1998;16:2879–2883.
Whang YE, Wu X, Suzuki H, et al. Inactivation of the adenocarcinoma suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA 1998;95:5246–5250.
Sarker D, Reid AH, Yap TA, et al. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res 2009;15:4799–4805.
Hernandez S, de Muga S, Agell L, et al. FGFR3 mutations in prostate cancer: association with low-grade adenocarcinomas. Mod Pathol 2009;22:848–856.
Cai CQ, Peng Y, Buckley MT, et al. Epidermal growth factor receptor activation in prostate cancer by three novel missense mutations. Oncogene 2008;27:3201–3210.
Abdel-Rahman MH, Yang Y, Zhou XP, et al. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol 2006;24:288–295.
Tashiro H, Blazes MS, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res 1997;57:3935–3940.
Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275:1943–1947.
Brandt B, Meyer-Staeckling S, Schmidt H, et al. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res 2006;12:7252–7260.
Yamamoto H, Toyooka S, Mitsudomi T . Impact of EGFR mutation analysis in non-small cell lung cancer. Lung Cancer 2009;63:315–321.
Zhang X, Chang A . Somatic mutations of the epidermal growth factor receptor and non-small-cell lung cancer. J Med Genet 2007;44:166–172.
Mitsudomi T, Yatabe Y . Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci 2007;98:1817–1824.
Shuch B, Mikhail M, Satagopan J, et al. Racial disparity of epidermal growth factor receptor expression in prostate cancer. J Clin Oncol 2004;22:4725–4729.
Zellweger T, Ninck C, Bloch M, et al. Expression patterns of potential therapeutic targets in prostate cancer. Int J Cancer 2005;113:619–628.
Ibrahim GK, Kerns BJ, MacDonald JA, et al. Differential immunoreactivity of epidermal growth factor receptor in benign, dysplastic and malignant prostatic tissues. J Urol 1993;149:170–173.
Morris GL, Dodd JG . Epidermal growth factor receptor mRNA levels in human prostatic adenocarcinomas and cell lines. J Urol 1990;143:1272–1274.
Turkeri LN, Sakr WA, Wykes SM, et al. Comparative analysis of epidermal growth factor receptor gene expression and protein product in benign, premalignant, and malignant prostate tissue. Prostate 1994;25:199–205.
Parsons R . Human cancer, PTEN and the PI-3 kinase pathway. Semin Cell Dev Biol 2004;15:171–176.
Uzoh CC, Perks CM, Bahl A, et al. PTEN-mediated pathways and their association with treatment-resistant prostate cancer. BJU Int 2009;104:556–561.
Feilotter HE, Nagai MA, Boag AH, et al. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene 1998;16:1743–1748.
McMenamin ME, Soung P, Perera S, et al. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res 1999;59:4291–4296.
Podsypanina K, Ellenson LH, Nemes A, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA 1999;96:1563–1568.
Ratnacaram CK, Teletin M, Jiang M, et al. Temporally controlled ablation of PTEN in adult mouse prostate epithelium generates a model of invasive prostatic adenocarcinoma. Proc Natl Acad Sci USA 2008;105:2521–2526.
Acknowledgements
The study was funded with grants FIS/Instituto Carlos III/FEDERPI06/1411 and PS09/01106 from the Spanish Ministry of Health and Support Grant 2008 from the Spanish Association Against Cancer (Barcelona Territorial Board). We thank Sergi Mojal (Department of Biostatistics, Institut Municipal d’Investigació Mèdica, Hospital del Mar, Barcelona, Spain) for the technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
de Muga, S., Hernández, S., Agell, L. et al. Molecular alterations of EGFR and PTEN in prostate cancer: association with high-grade and advanced-stage carcinomas. Mod Pathol 23, 703–712 (2010). https://doi.org/10.1038/modpathol.2010.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.45
Keywords
This article is cited by
-
Loss of miR-936 leads to acquisition of androgen-independent metastatic phenotype in prostate cancer
Scientific Reports (2022)
-
An integrated genomic and metabolomic approach for defining survival time in adult oligodendrogliomas patients
Metabolomics (2019)
-
PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: implications for active surveillance
Modern Pathology (2016)
-
Prostate Cancer in African American Men: The Effect of Androgens and microRNAs on Epidermal Growth Factor Signaling
Hormones and Cancer (2016)
-
Emerging roles of radioresistance in prostate cancer metastasis and radiation therapy
Cancer and Metastasis Reviews (2014)