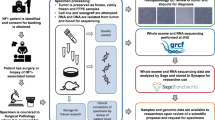
Abstract
Neurothekeomas and dermal nerve sheath myxomas have previously been considered related cutaneous neoplasms of peripheral nerve sheath origin based on light microscopic similarities. However, recent immunohistochemical and ultrastructural data indicate nerve sheath myxomas exhibit true nerve sheath differentiation, whereas no such compelling evidence exists for neurothekeomas. Although neurothekeomas lack a specific immunohistochemical profile, similar antigen expression and histopathologic patterns suggest neurothekeomas may be categorized as fibrohistiocytic tumors. To date, no known molecular studies have examined the histogenetic relationship of these tumors. We report the first microarray-based gene expression profile study of these entities on formalin-fixed paraffin-embedded tissues. Cases of dermal schwannomas, dermal nerve sheath myxomas, myxoid/mixed/cellular neurothekeomas, and cellular fibrous histiocytomas diagnosed in the past 3 years were identified in our database. Archival formalin-fixed paraffin-embedded tissue from 28 patients was selected for microarray analysis (seven schwannomas, five nerve sheath myxomas, nine myxoid/mixed/cellular neurothekeomas and seven cellular fibrous histiocytomas). Following tumor RNA isolation, amplification and labeling using commercially available kits, labeled targets were hybridized to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Santa Clara, CA, USA). Acquisition of array images and data analyses was performed using appropriate software. Hierarchical clustering and principal component analysis demonstrated discrete groups, which correlated with histopathologically identified diagnoses. Dermal nerve sheath myxomas demonstrate very similar molecular genetic signatures to dermal schwannomas, whereas neurothekeomas of all subtypes more closely resemble cellular fibrous histiocytomas. We are the first to report distinct gene expression profiles for nerve sheath myxomas and neurothekeomas, which further substantiates the argument that these are separate entities. Our molecular data confirms that dermal nerve sheath myxomas are of peripheral nerve sheath origin, and suggests that neurothekeomas may actually be a variant of fibrous histiocytomas.
Similar content being viewed by others
Main
Gallager and Helwig1 initially described neurothekeomas in 1980 as benign cutaneous tumors of nerve sheath differentiation. Subsequent authors suggested neurothekeomas represented variants of dermal nerve sheath myxomas,2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 which were initially described in 1969 by Harkin and Reed.13 The presumed relationship between neurothekeomas and nerve sheath myxomas was inferred from histopathologic similarities between these two entities and the resemblance of neurothekeomas to non-neoplastic neural structures. Additionally, some authors demonstrated that the immunohistochemical profile of neurothekeomas was compatible with cultured cells of nerve sheath origin from rats, which suggested that neurothekeomas represented cells of immature nerve sheath differentiation.14, 15, 16, 17
However, subtle morphologic differences, divergent immunohistochemical staining in human tissue and ultrastructural studies led many authors to question the relationship between neurothekeomas and nerve sheath myxomas.1, 7, 9, 11, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Recent work by Fetsch et al,28 using a combination of morphological, immunohistochemical and ultrastructural data, strongly argues that nerve sheath myxomas represent a myxoid variant of schwannoma. Using similar studies, Hornick and Fetsch both underscore previous suggestions that neurothekeomas represent a variant of cutaneous fibrohistiocytic tumors, which are benign fibroblastic and histiocytic tumors with a wide range of histomorphologic appearances.27, 29
To date, there have been no molecular studies examining these entities. Recent biotechnological advances have optimized conditions to improve the quality and quantity of RNA extracted from formalin-fixed paraffin-embedded tissues.30, 31, 32 These now permit molecular analysis, including microarray-based whole-genome-wide testing, of clinically small lesions from archival tissues.
We report the first microarray-based gene expression profile study of neurothekeomas, nerve sheath myxomas, schwannomas and cellular fibrous histiocytomas. We successfully used formalin-fixed paraffin-embedded material from archived diagnostic tissue. Our data further clarifies the relationship between these entities and demonstrates that clinically significant gene expression profiles can be obtained from routine biopsy material.
Materials and methods
A search for dermatopathology cases coded as ‘neurothekeoma’, ‘nerve sheath myxoma’, ‘cellular fibrohistiocytoma’ and ‘schwannoma’ was performed on the University of California, Los Angeles (UCLA), Department of Pathology database. The search was limited to cases from the previous 3 years, as early literature indicated that this period of storage did not adversely affect RNA extraction for microarray analysis in formalin-fixed paraffin-embedded blocks.30, 33, 34
All slides were re-reviewed by three authors (SS, SB and SD) and appropriately categorized based on histological and immunohistochemical criteria set forth by the Armed Forces Institute of Pathology, World Health Organization and Hornick and Fletcher's reviews of neurothekeomas and cellular fibrohistiocytic tumors (Table 1).22, 27, 28, 29, 35, 36 Exclusion criteria included cases with scant tissue remaining in the block and cases without unanimous agreement on the definitive diagnosis. Analysis was also limited to blocks of tissue with at least 40% tumor cells. In all, 28 cases were selected from Department files or retrievable from originating hospitals for cases seen in consult. These cases included patients diagnosed with myxoid, mixed or cellular neurothekeomas (nine cases), dermal nerve sheath myxomas (five cases), cellular fibrous histiocytomas (seven cases) and dermal schwannomas (seven cases; Figures 1, 2, 3, 4). Areas of the slide (∼5 mm2) with maximal tumor tissue were circled and corresponding areas were macrodissected from the tumor blocks for RNA isolation.
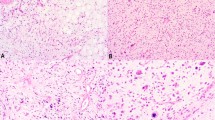
(a) Cutaneous cellular fibrous histiocytoma (BFH no. 2) centered within the dermis with overlying epidermal hyperplasia (H and E stain; × 20). (b) Cellular fibrous histiocytoma (BFH no. 5) with typical dermal collagen entrapment and haphazard cellular pattern (H and E stain; × 28). (c) Positive staining for factor 13a in BFH no. 5 (factor 13A; × 200).
(a) Myxoid neurothekeoma (MyxNK no. 3) centered within the dermis with broad fibrous septae (H and E; × 20). (b) Higher power showing epithelioid cytomorphology within a prominent myxoid matrix (H and E; × 200). (c) Cellular neurothekeoma (CNK no. 1) with a plexiform, whorled pattern of cells within the dermis (H and E; × 60). (d) Cellular neurothekeoma (CNK no. 6) composed of plump epithelioid cells in a vaguely whorled pattern and faintly myxoid matrix (H and E; × 100).
(a) Multilobulated and circumscribed dermal nerve sheath myxoma (NSM no. 3) with prominent fibrous bands (H and E; × 10). (b) Higher power demonstrates prominent stellate to spindled cells within abundant myxoid matrix (H and E; × 28). (c) Acral skin with a nerve sheath myxoma (NSM no. 1) demonstrating a plexiform pattern of paucicellular, spindled cells within a myxoid matrix (H and E; × 40). (d) Strong-positive staining of stellate to spindled cells with S100 protein (S100; × 40).
A variety of histopathologic data were collected, including the presence of epidermis/adnexal structures, degree of inflammation and percent of tumor/fibrous/dermal/adipose tissue in the areas of macrodissection (Table 2). Immunoperoxidase studies using the avidin–biotin–peroxidase technique performed included S100 protein (Ventana, Tucson, AZ, USA; predilute), α-smooth muscle actin (Ventana; predilute), factor XIIIa (Calbiochem, Merck & Co., Whitehouse Station, NJ, USA; 1:30), neuron-specific enolase (Ventana; predilute), and NKI/C3 (BioGenex, Fremont, CA, USA; 1:1600).
All RNA extraction and microarray experiments were performed at the UCLA Department of Pathology Clinical Microarray Core Laboratory under the direction of one of the authors (XL). Total RNA was isolated using the Ambion RecoverAll (Applied Biosystems/Ambion, Austin, TX, USA) kit according to the manufacturer's instructions. RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and purity/concentration was determined using a NanoDrop 8000 (NanoDrop Products, Wilmington, DE, USA). Microarray targets were prepared using NuGEN WT-Ovation Formalin-Fixed Paraffin-Embedded RNA Amplification System and FL-Ovation cDNA Biotin Module V2 (NuGEN Technologies, San Carlos, CA, USA) and then hybridized to the Affymetrix GeneChip U133 Plus 2.0 Array (Affymetrix), all according to manufacturers' instructions. The arrays were washed and stained with streptavidin phycoerythrin in Affymetrix Fluidics Station 450 using the Affymetrix GeneChip protocol, and then scanned using an Affymetrix GeneChip Scanner 3000.
The acquisition and initial quantification of array images were conducted using the AGCC software (Affymetrix). The subsequent data analyses were performed using Partek Genomics Suite Version 6.4 (Partek, St Louis, MO, USA). Differentially expressed genes were selected at ≥threefold and P<0.005. Cluster analysis and principal component analysis were conducted with Partek default settings. Biofunctional analysis was performed using Ingenuity Pathways Analysis Software Version 7.6 (Ingenuity Systems, Redwood City, CA, USA).
Results
The clinical and immunohistochemical features of our cases are similar to those recently described in the literature (Tables 3 and 4).27, 28, 29
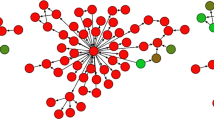
The gene expression profiles of these four histological entities are shown in Figures 5, 6, 7. Greater numbers of differentially expressed genes between nerve sheath myxomas and neurothekeomas suggest that they are not related (Figure 5). This data, in conjunction with cluster analysis (Figure 6) and principal component analysis (Figure 7), demonstrate two molecular groups: a nerve sheath myxoma/schwannoma group and a neurothekeoma/cellular fibrous histiocytoma group.
Cluster map constructed using 452 differentially expressed genes between neurothekeomas and nerve sheath myxomas. The nerve sheath myxomas and schwannomas are clustered into one group, whereas the neurothekeomas and cellular fibrous histiocytomas fall into another group (>threefold change; P-value <0.005).
Principal component analysis using 452 differentially expressed genes between neurothekeomas and nerve sheath myxomas demonstrates one major group composed of schwannomas and nerve sheath myxomas and another major group composed of neurothekeomas and cellular fibrous histiocytomas (>threefold change; P-value <0.005).
Genes upregulated in nerve sheath myxomas and schwannomas compared with both neurothekeomas and cellular fibrous histiocytomas primarily included those involved in peripheral nerve sheath maintenance or development (Table 5), with the most statistically significant differentially gene expressed being S100B.37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48 The principal differentially expressed genes upregulated in neurothekeomas and cellular fibrous histiocytomas over nerve sheath myxomas and schwannomas consisted of those encoding various metalloproteinases and glycoproteins. These genes are involved in extracellular matrix growth and remodeling, and cell adhesion in fibroblasts and macrophages (Table 6).49, 50, 51, 52, 53, 54, 55, 56
There were relatively few statistically significant genes differentially expressed between neurothekeomas and cellular fibrous histiocyomas, and between nerve sheath myxomas and schwannomas. For neurothekeomas and cellular fibrous histiocytomas, these genes primarily included various collagen proteins. Between nerve sheath myxomas and schwannomas, differentially expressed genes included those encoding neuronal cell intercellular signaling, collagen and cell adhesion molecules.
One outlier (nerve sheath myxoma no. 1) did not cluster with the schwannomas/nerve sheath myxomas group, despite typical histological and immunohistochemical features. Upon analysis of the cluster map, this sample demonstrated broadly decreased expression of almost all genes compared with the other samples (Figure 6). Outside processing and storage of the block, technical limitations/artifact inherent to microarray analysis of formalin-fixed paraffin-embedded tissue or a higher proportion of fibrous tissue in this case (60% vs an overall average of 35% in the other cases) may account for this outcome (Table 2).
Discussion
Whole-genome microarray and transcription profiling analysis simultaneously interrogates tens of thousands of genes, and thus provides sophisticated molecular images of tissues. However, until recently, this technology could not be applied to formalin-fixed paraffin-embedded tissue blocks. Formalin fixation causes chemical cross-linking of nucleic acids to each other and to proteins, as well as fragmentation of nucleic acids. Paraffin embedding results in further mechanical stresses to RNA. Cumulatively, these lead to suboptimal nucleic acid quality and hampered reverse transcription and amplification reactions.57, 58, 59, 60 Thus, in the past, gene expression profile studies were restricted to tissues with higher intrinsic nucleic acid quality, such as cell lines, fresh blood or fresh frozen tissue.61, 62, 63, 64, 65, 66
Recent biotechnological advances have improved RNA isolation and amplification techniques, resulting in successful use of formalin-fixed paraffin-embedded tissues to identify gene signatures in human tumors. These advances include newer formalin-fixed paraffin-embedded tissue extraction protocols, cDNA-mediated annealing, selection, extension and ligation, the addition of random hexamer priming to oligo(dT) priming and newly developed array platforms with redesigned probes.30, 31, 32, 33, 34, 57, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82 Although sensitivity is low, high specificity and positive predictive value suggest that transcript detection is reliable from formalin-fixed paraffin-embedded tissue.33
Although RNA isolation techniques from formalin-fixed paraffin-embedded tissues have improved, problems remain, largely because of the greater fragmented nature of RNA in formalin-fixed paraffin-embedded tissue.30, 32, 75, 78 Magnitudes of differential expression are often higher in formalin-fixed paraffin-embedded tissue than in fresh frozen samples, because of the increased background noise; this increases the odds of false-positive results.30, 83 Despite these concerns, several studies have shown comparable biological information between genome-wide microarray analysis from matched fresh frozen and formalin-fixed paraffin-embedded tissue.30, 32, 72, 74, 75, 77, 78
The use of formalin-fixed paraffin-embedded rather than fresh frozen tissue clearly allows more practical applications of gene microarray analysis. The National Cancer Institute estimates that 85% of all cancer surgeries are performed in community hospitals, which historically have not had the resources for procuring snap frozen tumor samples.84 Small tumors often must be completely paraffin embedded for diagnostic studies. In some cases, the diagnosis is not apparent until microscopic examination, at which time fresh tissue is unavailable. Formalin-fixed paraffin-embedded tissue allows for improved correlation between histopathologic findings and molecular signatures, allowing for better understanding of gene expression profiles. Finally, use of archival formalin-fixed paraffin-embedded tissue allows for large-scale retrospective studies; as clinical and outcome data can be associated with these tissues, this permits more clinically relevant research.
We are the first to report gene expression profiles for nerve sheath myxomas, schwannomas, neurothekeomas and cellular fibrous histiocytomas. Similar grouping of schwannomas and nerve sheath myxomas by principal component analysis and hierarchical clustering strongly suggests that these lesions are closely related. Functional gene analysis of the microarray profiles revealed that many of the highest differentially expressed genes in schwannomas/nerve sheath myxomas compared with neurothekeomas/cellular fibrous histiocytomas involve peripheral nerve sheath development and maintenance. This data strongly supports previous histomorphologic studies, indicating that nerve sheath myxomas and schwannomas are related neoplasms of peripheral nerve sheath origin and, despite some similar histological features, are unrelated to neurothekeomas.27, 28, 29
Neurothekeomas expressed genes that primarily encoded glycoproteins and metalloproteinases, often from macrophages and fibroblasts involved in matrix growth and remodeling, angiogenesis and mesenchymal cell differentiation.49, 50, 51, 52, 53, 54, 55, 56 The S100B gene was the most significantly differentially expressed gene between nerve sheath myxomas and neurothekeomas. This further provides molecular support for recent work reporting negative S100 protein staining by immunohistochemistry in neurothekeomas of all subtypes.27, 29 Interestingly, our microarray data demonstrated increased podoplanin gene expression in neurothekeomas over nerve sheath myxomas. This molecular finding correlates with recent immunohistochemical observations that podoplanin (D2-40) protein expression is seen in neurothekeomas.85
Our gene expression profile data for neurothekeomas and cellular fibrous histiocytomas support previous arguments that these are related neoplasms.1, 7, 9, 11, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 29, 86 Both groups showed similar expression of genes involved in matrix remodeling, and growth involving fibroblasts and histiocytes. Analysis of the differential gene expression data between neurothekeomas and cellular fibrous histiocytomas was less instructive. The vast majority of the top differentially expressed genes between these tumors encoded various uncharacterized loci or nonspecific immunoglobulin loci, zinc fingers, chemokine receptors or proteins involved in intracellular transport. There is no definite known relationship between these gene products and cellular fibrous histiocytomas in the literature. Thus, the significance of these findings is unclear. A small number of differences in expression of genes involved in connective tissue matrix and tissue remodeling were noted, and it is possible that this is secondary to the inclusion of both myxoid and mixed types of neurothekeomas.
Our data does not resolve the question of whether neurothekeomas are simply dermal variants of the rarer plexiform variant of fibrohistiocytic tumors. Neurothekeomas and plexiform fibrous histiocytomas share nearly identical demographic and anatomic distributions, as well as many overlapping histological features.7, 27, 29, 87, 88, 89 There are no reliable immunohistochemical markers that distinguish these lesions.29, 89 The majority of plexiform fibrous histiocytomas are clinically benign, similar to neurothekeomas. Taken together, all these features have led some authors to suggest that neurothekeomas and plexiform fibrohistiocytomas are the same lesion.89, 90 It has been suggested that the deeper subcutaneous location of plexiform fibrous histiocytomas is responsible for the higher rates of local recurrence and rare instances of metastases.24, 87, 88, 89, 90 With one exception, the cases of plexiform fibrous histiocytomas seen at our institution during the study were outside consultations for which we were not able to obtain material for testing. Molecular profiling by gene microarray analysis would be a critically helpful adjunct in determining the relationship between these entities.
Our molecular data provides critical insights into the relationship between these entities. However, practically, diagnosis of these entities will rely on light microscopy and immunohistochemistry for the present time. Gene microarray studies are expensive compared with standard pathology techniques. Recent large histological and immunohistochemical studies of these entities have substantially clarified the microscopic features of each.27, 28, 29 Furthermore, the clinical implications of misdiagnosis are minimal, as the vast majority of these are clinically benign. Collectively, these features do not support use of very expensive microarray technology for routine diagnosis.
In summary, our gene expression array data strongly supports that nerve sheath myxomas are of peripheral nerve sheath origin and are distinct neoplasms from neurothekeomas. Furthermore, our data shows many similarities in gene expression between neurothekeomas and cellular fibrous histiocytomas, supporting recent arguments that these entities may in fact be related. Further studies to examine the molecular profile of fibrous histiocytomas and neurothekeomas are indicated, in particular the rare plexiform variant of fibrous histiocytomas, which shows many histological and clinical similarities to neurothekeomas. Additionally, our data shows that advances in RNA isolation, amplification, and labeling protocols allow successful gene microarray analysis on routine clinical biopsy samples that are processed as formalin-fixed paraffin-embedded tissues.
References
Gallager RL, Helwig EB . Neurothekeoma—a benign cutaneous tumor of neural origin. Am J Clin Pathol 1980;74:759–764.
Pulitzer DR, Reed RJ . Nerve-sheath myxoma (perineurial myxoma). Am J Dermatopathol 1985;7:409–421.
Allen PW . Myxoma is not a single entity: a review of the concept of myxoma. Ann Diagn Pathol 2000;4:99–123.
Argenyi ZB, Balogh K, Abraham AA . Degenerative (‘ancient’) changes in benign cutaneous schwannoma. A light microscopic, histochemical and immunohistochemical study. J Cutan Pathol 1993;20:148–153.
Barnhill RL, Mihm Jr MC . Cellular neurothekeoma A distinctive variant of neurothekeoma mimicking nevomelanocytic tumors. Am J Surg Pathol 1990;14:113–120.
Barnhill RL, Dickersin GR, Nickeleit V, et al. Studies on the cellular origin of neurothekeoma: clinical, light microscopic, immunohistochemical, and ultrastructural observations. J Am Acad Dermatol 1991;25:80–88.
Hollowood K, Holley MP, Fletcher CD . Plexiform fibrohistiocytic tumour: clinicopathological, immunohistochemical and ultrastructural analysis in favour of a myofibroblastic lesion. Histopathology 1991;19:503–513.
Bhaskar AR, Kanvinde R . Neurothekeoma of the hand. J Hand Surg Br 1999;24:631–633.
Fullen DR, Lowe L, Su LD . Antibody to S100a6 protein is a sensitive immunohistochemical marker for neurothekeoma. J Cutan Pathol 2003;30:118–122.
Husain S, Silvers DN, Halperin AJ, et al. Histologic spectrum of neurothekeoma and the value of immunoperoxidase staining for S-100 protein in distinguishing it from melanoma. Am J Dermatopathol 1994;16:496–503.
Rosati LA, Fratamico FC, Eusebi V . Cellular neurothekeoma. Appl Pathol 1986;4:186–191.
Connolly M, Hickey JR, Intzedy L, et al. Subungual neurothekeoma. J Am Acad Dermatol 2005;52:159–162.
Harkin JC, Reed RJ . Tumors of the peripheral nervous system. In: Rosai J (ed). Atlas of Tumor Pathology Vol. 2nd series Fascicle 3, Armed Forces Institute of Pathology: Washington DC,, 1969, pp 29–65.
Vougioukas VI, Roeske S, Michel U, et al. Wallerian degeneration in ICAM-1-deficient mice. Am J Pathol 1998;152:241–249.
Yamate J, Yasui H, Benn SJ, et al. Characterization of newly established tumor lines from a spontaneous malignant schwannoma in F344 rats: nerve growth factor production, growth inhibition by transforming growth factor-beta1, and macrophage-like phenotype expression. Acta Neuropathol 2003;106:221–233.
Misago N, Satoh T, Narisawa Y . Cellular neurothekeoma with histiocytic differentiation. J Cutan Pathol 2004;31:568–572.
Griffin JW, George R, Ho T . Macrophage systems in peripheral nerves. A review. J Neuropathol Exp Neurol 1993;52:553–560.
Angervall L, Kindblom LG, Haglid K . Dermal nerve sheath myxoma. A light and electron microscopic, histochemical and immunohistochemical study. Cancer 1984;53:1752–1759.
Blumberg AK, Kay S, Adelaar RS . Nerve sheath myxoma of digital nerve. Cancer 1989;63:1215–1218.
Wang AR, May D, Bourne P, et al. PGP9.5: a marker for cellular neurothekeoma. Am J Surg Pathol 1999;23:1401–1407.
Laskin WB, Fetsch JF, Miettinen M . The ‘neurothekeoma’: immunohistochemical analysis distinguishes the true nerve sheath myxoma from its mimics. Hum Pathol 2000;31:1230–1241.
Scheithauer B, Woodruff J, Erlandson R . Miscellaneous benign neurogenic tumors. In: Rosai J, Sobin L (eds). Atlas of Tumor Pathology Tumors of the Peripheral Nervous System, 3rd series Armed Forces Institute of Pathology: Washington DC, 1999, pp 219–282.
Zelger BG, Steiner H, Kutzner H, et al. Cellular ‘neurothekeoma’: an epithelioid variant of dermatofibroma? Histopathology 1998;32:414–422.
Salomao DR, Nascimento AG . Plexiform fibrohistiocytic tumor with systemic metastases: a case report. Am J Surg Pathol 1997;21:469–476.
Requena L, Sangueza OP . Benign neoplasms with neural differentiation: a review. Am J Dermatopathol 1995;17:75–96.
Paulus W, Jellinger K, Perneczky G . Intraspinal neurothekeoma (nerve sheath myxoma). A report of two cases. Am J Clin Pathol 1991;95:511–516.
Fetsch JF, Laskin WB, Hallman JR, et al. Neurothekeoma: an analysis of 178 tumors with detailed immunohistochemical data and long-term patient follow-up information. Am J Surg Pathol 2007;31:1103–1114.
Fetsch JF, Laskin WB, Miettinen M . Nerve sheath myxoma: a clinicopathologic and immunohistochemical analysis of 57 morphologically distinctive, S-100 protein- and GFAP-positive, myxoid peripheral nerve sheath tumors with a predilection for the extremities and a high local recurrence rate. Am J Surg Pathol 2005;29:1615–1624.
Hornick JL, Fletcher CD . Cellular neurothekeoma: detailed characterization in a series of 133 cases. Am J Surg Pathol 2007;31:329–340.
Fedorowicz G, Guerrero S, Wu TD, et al. Microarray analysis of RNA extracted from formalin-fixed, paraffin-embedded and matched fresh-frozen ovarian adenocarcinomas. BMC Med Genomics 2009;2:23.
Koh SS, Opel ML, Wei JP, et al. Molecular classification of melanomas and nevi using gene expression microarray signatures and formalin-fixed and paraffin-embedded tissue. Mod Pathol 2009;22:538–546.
Ravo M, Mutarelli M, Ferraro L, et al. Quantitative expression profiling of highly degraded RNA from formalin-fixed, paraffin-embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest 2008;88:430–440.
Linton KM, Hey Y, Saunders E, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer 2008;98:1403–1414.
Paik S, Kim CY, Song YK, et al. Technology insight: application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Clin Pract Oncol 2005;2:246–254.
LeBoit PE, Burg G, Weedon D, et al. Pathology and Genetics of Skin Tumours. IARC Press: Lyon, 2006.
Calonje E, Mentzel T, Fletcher CD . Cellular benign fibrous histiocytoma. Clinicopathologic analysis of 74 cases of a distinctive variant of cutaneous fibrous histiocytoma with frequent recurrence. Am J Surg Pathol 1994;18:668–676.
Scotto C, Deloulme JC, Rousseau D, et al. Calcium and S100B regulation of p53-dependent cell growth arrest and apoptosis. Mol Cell Biol 1998;18:4272–4281.
Cheng Y, Cheung M, Abu-Elmagd MM, et al. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res 2000;121:233–241.
Giese KP, Martini R, Lemke G, et al. Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell 1992;71:565–576.
Schaich M, Budzinski RM, Stoffel W . Cloned proteolipid protein and myelin basic protein cDNA transcription of the two genes during myelination. Biol Chem Hoppe Seyler 1986;367:825–834.
Mandai K, Nakanishi H, Satoh A, et al. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J Cell Biol 1999;144:1001–1017.
Vits L, Van Camp G, Coucke P, et al. MASA syndrome is due to mutations in the neural cell adhesion gene L1CAM. Nat Genet 1994;7:408–413.
Harroch S, Furtado GC, Brueck W, et al. A critical role for the protein tyrosine phosphatase receptor type Z in functional recovery from demyelinating lesions. Nat Genet 2002;32:411–414.
Wakamatsu Y, Endo Y, Osumi N, et al. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Dev Dyn 2004;229:74–86.
Narayanan V, Ripepi B, Jabs EW, et al. Partial structure and mapping of the human myelin P2 protein gene. J Neurochem 1994;63:2010–2013.
Miyoshi K, Honda A, Baba K, et al. Disrupted-in-schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry 2003;8:685–694.
Lodge AP, McNamee CJ, Howard MR, et al. Identification and characterization of CEPU-Se-A secreted isoform of the IgLON family protein, CEPU-1. Mol Cell Neurosci 2001;17:746–760.
Arai M, Itokawa M, Yamada K, et al. Association of neural cell adhesion molecule 1 gene polymorphisms with bipolar affective disorder in Japanese individuals. Biol Psychiatry 2004;55:804–810.
Kurada P, White K . Epidermal growth factor receptor: its role in Drosophila eye differentiation and cell survival. Apoptosis 1999;4:239–243.
Lafuste P, Sonnet C, Chazaud B, et al. ADAM12 and alpha9beta1 integrin are instrumental in human myogenic cell differentiation. Mol Biol Cell 2005;16:861–870.
Boire A, Covic L, Agarwal A, et al. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 2005;120:303–313.
Shao R, Bao S, Bai X, et al. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol 2004;24:3992–4003.
Tsukahara S, Ikeda R, Goto S, et al. Tumour necrosis factor alpha-stimulated gene-6 inhibits osteoblastic differentiation of human mesenchymal stem cells induced by osteogenic differentiation medium and BMP-2. Biochem J 2006;398:595–603.
Kos J, Sekirnik A, Premzl A, et al. Carboxypeptidases cathepsins X and B display distinct protein profile in human cells and tissues. Exp Cell Res 2005;306:103–113.
Aertgeerts K, Levin I, Shi L, et al. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J Biol Chem 2005;280:19441–19444.
Schacht V, Dadras SS, Johnson LA, et al. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. Am J Pathol 2005;166:913–921.
Masuda N, Ohnishi T, Kawamoto S, et al. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 1999;27:4436–4443.
Penland SK, Keku TO, Torrice C, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest 2007;87:383–391.
Karsten SL, Van Deerlin VM, Sabatti C, et al. An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res 2002;30:E4.
Benchekroun M, DeGraw J, Gao J, et al. Impact of fixative on recovery of mRNA from paraffin-embedded tissue. Diagn Mol Pathol 2004;13:116–125.
Hoheisel JD . Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet 2006;7:200–210.
Sotiriou C, Piccart MJ . Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer 2007;7:545–553.
Brazma A, Vilo J . Gene expression data analysis. Microbes Infect 2001;3:823–829.
Frueh FW . Impact of microarray data quality on genomic data submissions to the FDA. Nat Biotechnol 2006;24:1105–1107.
Farragher SM, Tanney A, Kennedy RD, et al. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem Cell Biol 2008;130:435–445.
Kunz M, Ibrahim SM, Koczan D, et al. DNA microarray technology and its applications in dermatology. Exp Dermatol 2004;13:593–606.
Hey Y, Pepper SD . Interesting times for microarray expression profiling. Brief Funct Genomic Proteomic 2009;8:170–173.
Loudig O, Milova E, Brandwein-Gensler M, et al. Molecular restoration of archived transcriptional profiles by complementary-template reverse-transcription (CT-RT). Nucleic Acids Res 2007;35:e94.
Xiang CC, Chen M, Ma L, et al. A new strategy to amplify degraded RNA from small tissue samples for microarray studies. Nucleic Acids Res 2003;31:e53.
Xiang Z, Yang Y, Ma X, et al. Microarray expression profiling: analysis and applications. Curr Opin Drug Discov Devel 2003;6:384–395.
Chen J, Byrne Jr GE, Lossos IS . Optimization of RNA extraction from formalin-fixed, paraffin-embedded lymphoid tissues. Diagn Mol Pathol 2007;16:61–72.
Bibikova M, Yeakley JM, Chudin E, et al. Gene expression profiles in formalin-fixed, paraffin-embedded tissues obtained with a novel assay for microarray analysis. Clin Chem 2004;50:2384–2386.
Bibikova M, Yeakley JM, Wang-Rodriguez J, et al. Quantitative expression profiling of RNA from formalin-fixed, paraffin-embedded tissues using randomly assembled bead arrays. Methods Mol Biol 2008;439:159–177.
Lee J, Hever A, Willhite D, et al. Effects of RNA degradation on gene expression analysis of human postmortem tissues. Faseb J 2005;19:1356–1358.
Frank M, Doring C, Metzler D, et al. Global gene expression profiling of formalin-fixed paraffin-embedded tumor samples: a comparison to snap-frozen material using oligonucleotide microarrays. Virchows Arch 2007;450:699–711.
Coudry RA, Meireles SI, Stoyanova R, et al. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn 2007;9:70–79.
Scicchitano MS, Dalmas DA, Bertiaux MA, et al. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem 2006;54:1229–1237.
Lassmann S, Kreutz C, Schoepflin A, et al. A novel approach for reliable microarray analysis of microdissected tumor cells from formalin-fixed and paraffin-embedded colorectal cancer resection specimens. J Mol Med 2009;87:211–224.
Chung CH, Parker JS, Ely K, et al. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-kappaB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res 2006;66:8210–8218.
Haque T, Faury D, Albrecht S, et al. Gene expression profiling from formalin-fixed paraffin-embedded tumors of pediatric glioblastoma. Clin Cancer Res 2007;13:6284–6292.
Onken MD, Worley LA, Ehlers JP, et al. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res 2004;64:7205–7209.
Rogerson L, Darby S, Jabbar T, et al. Application of transcript profiling in formalin-fixed paraffin-embedded diagnostic prostate cancer needle biopsies. BJU Int 2008;102:364–370.
Van Deerlin VM, Gill LH, Nelson PT . Optimizing gene expression analysis in archival brain tissue. Neurochem Res 2002;27:993–1003.
Compton CC (ed). Introductory session. Proceedings of the 3rd Annual Biospecimen Research Network Symposium, 24–25 March, United States and Canadian Academy of Pathology 99th Annual Meeting, Washington, DC, 2010.
Kaddu S, Leinweber B . Podoplanin expression in fibrous histiocytomas and cellular neurothekeomas. Am J Dermatopathol 2009;31:137–139.
Fullen DR, Reed JA, Finnerty B, et al. S100A6 expression in fibrohistiocytic lesions. J Cutan Pathol 2001;28:229–234.
Enzinger FM, Zhang RY . Plexiform fibrohistiocytic tumor presenting in children and young adults. An analysis of 65 cases. Am J Surg Pathol 1988;12:818–826.
Remstein ED, Arndt CA, Nascimento AG . Plexiform fibrohistiocytic tumor: clinicopathologic analysis of 22 cases. Am J Surg Pathol 1999;23:662–670.
Jaffer S, Ambrosini-Spaltro A, Mancini AM, et al. Neurothekeoma and plexiform fibrohistiocytic tumor: mere histologic resemblance or histogenetic relationship? Am J Surg Pathol 2009;33:905–913.
Moosavi C, Jha P, Fanburg-Smith JC . An update on plexiform fibrohistiocytic tumor and addition of 66 new cases from the Armed Forces Institute of Pathology, in honor of Franz M Enzinger, MD. Ann Diagn Pathol 2007;11:313–319.
Acknowledgements
We thank our colleagues Maria Aguirre, Jian-Ping Cai, Frank Luo, Rebecca Remmers, and Celeste Vardaman, at Dianon Systems/LabCorp for contributing cases to this study. We also express our appreciation to the staff at the UCLA Translational Pathology Core Laboratory for providing digital image scanning of slides, the UCLA Pathology Outreach Services Office for retrieving slides and blocks, and William Schram for assistance in manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Abstract presented at the United States and Canadian Academy of Pathology 99th Annual Meeting, Washington, DC, 2010.
Rights and permissions
About this article
Cite this article
Sheth, S., Li, X., Binder, S. et al. Differential gene expression profiles of neurothekeomas and nerve sheath myxomas by microarray analysis. Mod Pathol 24, 343–354 (2011). https://doi.org/10.1038/modpathol.2010.203
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.203