Abstract
Designed ankyrin repeat proteins are a novel class of specific binding molecules, which display increased thermodynamic stability, smaller size and at least equal target affinity compared to immunoglobulins, making them potentially powerful tools in diagnostic pathology and therapeutic oncology. Here, we investigated whether designed ankyrin repeat proteins can reliably identify the amplification status of the epidermal growth factor receptor 2 in breast cancer. Designed ankyrin repeat proteins specific for epidermal growth factor receptor 2 were tested in paraffin-embedded tissue sections. Detection using enzymatic biotinylation proved to be most specific and sensitive. The affinity of the designed ankyrin repeat proteins was found crucial, but for a picomolar binder no further gain was found by making it multivalent. The best designed ankyrin repeat protein, G3 (KD 90 pM) was compared on breast cancer tissue microarrays (n=792) to an FDA-approved rabbit monoclonal antibody against epidermal growth factor receptor 2 (clone 4B5; Ventana Medical Systems) and correlated with corresponding epidermal growth factor receptor 2 amplification status measured by fluorescent in situ hybridization. Amplification status and epidermal growth factor receptor 2 expression measured by designed ankyrin repeat protein and antibody correlated strongly with each other (P<0.0001 each), the correlation between designed ankyrin repeat protein and amplification status being the strongest (0.87 compared to 0.77 for the antibody, Kendall's tau-beta). Using a modified scoring system for the designed ankyrin repeat protein, we show that the designed ankyrin repeat protein detects a positive epidermal growth factor receptor 2 amplification status with similar sensitivity and significantly higher specificity than the antibody (P=0.0005). This study suggests that designed ankyrin repeat proteins provide a valuable alternative to antibodies for the detection of epidermal growth factor receptor 2 expression in breast cancer and adds further compelling evidence for the use of designed ankyrin repeat proteins in diagnostic pathology and therapeutic oncology.
Similar content being viewed by others
Introduction
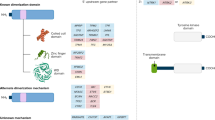
The human epidermal growth factor receptor 2 gene (HER2; Figure 1a) is overexpressed in a subset of breast carcinomas.1 Gene amplification is the primary mechanism of HER2 overexpression and is not only a predictor of poor prognosis but also indicates therapeutic options. Among them, trastuzumab, a monoclonal antibody that targets HER2 is effective in combination therapy with chemotherapeutic agents in both metastatic and adjuvant settings.2, 3, 4, 5 It improves the prognosis of HER2 overexpressing breast cancers to those not overexpressing HER2, but is not effective on the latter.6 Therefore, the high cost and potential drawbacks of such therapeutic interventions demand accurate HER2 testing. For this purpose, HER2 status is routinely being assessed in tissue biopsies of breast cancer patients either by immunohistochemistry or fluorescent in situ hybridization.1, 7, 8, 9, 10, 11, 12, 13 Typically, expression is first determined by immunohistochemistry. In cases were immunohistochemistry results are equivocal, the amplification status is being analyzed by fluorescent in situ hybridization, because it represents the current gold standard to decide whether a patient may benefit from trastuzumab treatment.1, 14 It would be highly desirable to further increase the reliability of the immunohistochemistry, and thus we decided to investigate alternative HER2 detection molecules, different from antibodies.
Comparison of the sizes of the HER2 receptor (a) with an antibody of the IgG type (b) and a designed ankyrin repeat protein (c–e), drawn to scale (description in details, see Supplementary Information).
Currently, immunohistochemistry diagnostics are still carried out with conventional monoclonal antibodies (Figure 1b). The generation and testing of new antibodies useful for diagnostics has traditionally been one of trial and error, with almost no impact from modern methods of protein engineering and evolution. Today, however, such recombinant and evolutionary methods can generate practically high-affinity reagents against any target, independent of its immunogenicity, direct the response against the desired epitope and distinguish between very similar molecules, and this can already be planned in the evolution strategy, thereby greatly expanding diagnostic options.15, 16, 17 Moreover, an almost limitless range of molecular formats becomes available, regarding valency, multispecific molecules and novel detection strategies.
Designed ankyrin repeat proteins (DARPins) are a promising novel class of synthetic non-immunoglobulin binding proteins that offer several advantages over conventional antibodies.18, 19, 20, 21, 22, 23 Their favorable molecular properties, such as small size and high stability, make them ideal tools for diagnostic and therapeutic oncology (Figure 1c). Their very high expression levels (100–200 mg/l in Escherichia coli shake flasks) make them very cost-effective to produce in bacteria. They can be rapidly selected from a library to specifically bind to practically any desired target16, 24, 25, 26, 27 and picomolar affinities can be reached, for example, as shown for the molecules used in this study. These in vitro selection strategies are also less time consuming compared to the generation of hybridomas.
In this study, we show that a DARPin performs robustly in scoring HER2 overexpression in patient samples. We specifically chose this well-characterized diagnostic example to be able to directly compare the validity of the results with the traditional approaches. However, many of the great advantages of DARPins, namely their wide range of modification and engineering options, have not yet been exploited to their full potential in this study.
Materials and methods
DARPins: Cloning, Expression and Purification
The DARPins were cloned as monomeric, dimeric and trimeric variants (see Supplementary Methods and Supplementary Table 1; Figure 1c–e). Monovalent DARPins H10-2-G327 (abbreviated here as G3) and H3-3-B626 were expressed in various formats for detection with different antibodies.
Patients and Tissues
Breast cancer tissue microarrays with more than 2000 primary breast cancers, recurrences, lymph node and hematogenous metastases have been constructed from formalin-fixed, paraffin-embedded tissue.28 The project has been approved by the local ethics committee (reference number StV 12-2005). For this study, tissue microarrays of primary breast cancers were included (n=792).
Immunohistochemistry and DARPin-Histochemistry of HER2
For HER2 immunohistochemistry, all tissue microarrays were analyzed with the Ventana Benchmark automated staining system (Ventana Medical Systems, Tucson, AZ, USA) using Ventana reagents for the entire procedure. For antigen retrieval, slides were heated with cell conditioning solution for 1 h (CC1; Tris-based buffer with slightly alkaline pH) using a standard protocol. We used the FDA-approved rabbit mAb clone 4B5 (Ventana Medical Systems) directed against the cytoplasmic domain of HER2 as primary antibody according to the manufacturer's recommendation. Detection was carried out using the UltraView-DAB kit (Ventana Medical Systems), based on conversion of diaminobenzidine to a dye with multimeric horseradish peroxidase (HRP) coupled to a biotinylated secondary anti-rabbit antibody.
For DARPin-histochemistry, we exploited the recombinant nature of the DARPins and directly enzymatically biotinylated them quantitatively and remote from the binding site at an engineered AviTag (Figure 1c). For antigen retrieval, slides were treated either with protease using the Bond Enzyme Pretreatment Kit (cat. no. AR9551) or by heating in Bond Epitope Retrieval Solution1/2 (cat. no. AR9961/AR9649). Blocking of endogenous biotin was achieved using the Endogenous Avidin/Biotin Blocking Kit (Zymed, San Francisco, CA, USA). All tissue microarrays were analyzed with the BondMax staining system (Vision BioSystems, acquired by Leica Microsystems). Detection was carried out using the Bond Intense R Detection kit, which relies on detection of biotinylated reagents with streptavidin-HRP conjugate, according to the manufacturer's recommendations.
HER2-Fluorescent In Situ Hybridization
The status of the HER2-neu proto-oncogene was assessed on tissue microarrays by fluorescence in situ hybridization using FDA-approved, direct fluorescently labeled DNA probes obtained from Pathvysion (Vysis; Abott AG Diagnostic Division, Baar, Switzerland).1
Histological Analysis of Immunohistochemistry, DARPin-Histochemistry and Fluorescent In Situ Hybridization and Statistics
The interpretation of the membranous staining obtained by immunohistochemistry and DARPin-histochemistry as well as the fluorescence signals by fluorescent in situ hybridization was carried out according to the recommendations of the ASCO and the CAP (see Supplementary Methods).1 For statistical analysis see Supplementary Methods.
Results
Establishment and Optimization of DARPin-Histochemistry
To develop a method allowing the sensitive and reproducible staining of bound DARPins to HER2 in paraffin-embedded tissue sections, we established the direct detection of biotin on the genetically fused AviTag (Figure 1c) with streptavidin-coupled HRP using 3,3′-diaminobenzidine as a substrate. Application of this detection method resulted in the expected specific membranous staining pattern that correlated with the location of HER2 (Figure 2a). In contrast to the previously described DARPin-histochemistry approach by Zahnd et al.26 based on the detection of a genetically fused hemagglutinin (HA)-tag, the novel automated method displayed a much higher specificity for HER2 with hardly any loss in signal sensitivity (Supplementary Figure 1). Detection of DARPins with anti-myc tag antibodies or anti-His tag antibodies was found inferior (see Supplementary Methods).
(a–c) Establishment DARPin-histochemistry using clone G3 on a test-tissue microarray containing 10 HER2-amplified carcinomas (red), 10 carcinomas equivocal for amplification (yellow) and 20 non-amplified tumors (green, all × 5 magnification). (d–f) Consecutive tissue sections of an HER2-amplified tumor incubated with different anti-HER2 DARPin constructs (affinities are displayed in parentheses, bar represents 40 μm).
Next we established the optimal conditions for DARPin-histochemistry on paraffin-embedded tissue using the biotinylated clone G3 on a test-tissue microarray including 40 breast cancer specimens (amplified n=10, equivocal n=10 and non-amplified n=20), whose status had been verified by fluorescent in situ hybridization (Figure 2). Detection was directly achieved by streptavidin-coupled HRP using 3,3′-diaminobenzidine as a substrate. Without additional signal amplification, at a concentration of 5 μg/ml the DARPin clone exhibited a slightly weaker staining pattern than the FDA-approved antibody that uses a secondary antibody to amplify the signal (Figure 2a). We thus tested the concentration dependence of the DARPin and found no further increase with increasing concentration, but instead that the DARPin staining properties were remarkably stable in a range of 1–25 μg/ml (Figure 2b). This indicates that even the lowest concentrations of DARPins are already saturating, and that the affinity is so high that all epitopes are covered under these conditions. Moreover, even at the highest concentrations, no unspecific binding was observed, in contrast to many antibodies, underlining the high specificity of binding. Sensitivity appears limited by the single biotin, recruiting a stoichiometric number of peroxidase–streptavidin fusion proteins to the cell, offering further engineering opportunities. Conventional signal amplification with an anti-biotin antibody followed with an HRP-polymer-coupled secondary antibody lead to some background staining (data not shown), and therefore the background-free direct detection was used throughout.
Staining by the DARPin was much more intense when antigen-retrieval was accomplished by enzymatic proteolysis rather than denaturing conditions (Figure 2c). This is consistent with the fact that G3 binds to the extracellular domain IV of HER2,26, 27 which is a compact disulfide-bonded domain, and thus the DARPin recognizes a structural epitope rather than a linear one.
Because HER2 molecules are present in proximity on the cell surface, it is reasonable to investigate whether multivalent detection molecules might be able to exploit the gain in avidity. Multivalent DARPins can be made conveniently by connecting them by a linker at the genetic level. Biotinylated mono-, bi- and trivalent DARPins (Figure 1c–e) of G3 were compared in DARPin-histochemistry in a concentration of 5 μg/ml. The bi- and trivalent DARPin exhibited a slightly weaker signal than the monovalent DARPin (Figure 2d) when used at the same weight/volume concentration. This can be explained by the high affinity of G3, because at this concentration (and even below, see Figure 2b) HER2 is already fully saturated with the monovalent DARPin, there is no further gain by avidity. Because in the bi- and trivalent DARPins, several DARPin domains must ‘share’ one biotin, the number of recruited HRP molecules is actually less than that for the monomer, reemphasizing that the limit of detection is given by the detection system, not by the DARPin.
Next, we investigated the question whether the high affinity of DARPin G3 is of direct benefit in the detection. For this purpose, DARPins that are point mutants of G3 (KD 90 pM), namely G3-D, G3-AVD and G3-HAVD, displaying lower affinities27 (ranging from 1 to 280 nM, Supplementary Table 1) but binding to the same epitope, were tested on paraffin-embedded tissue sections. When comparing these DARPins in DARPin-histochemistry, a continuous decrease in signal intensity was observed with decreasing affinity (Figure 2e). We also tested the DARPin H6-3-B3 with an affinity of 28 nM to HER2. This DARPin had been previously shown to specifically stain HER2-overexpressing cancer tissue using a genetically fused HA tag for detection.26 Under the present more stringent conditions, it failed to stain HER2 in DARPin-histochemistry (Figure 2f and Supplementary Figure 2), consistent with the data of the G3 mutants in respect of affinity. Nevertheless, an HER2-specific membranous staining could be detected when the concentration of DARPin H6-3-B3 was increased by 10-fold (Supplementary Figure 2).
We then investigated if DARPins that recognize different epitopes in domain IV of the ECD of HER2 (Supplementary Table 1) might exhibit similar or better staining properties on paraffin-embedded tissues. Under the same conditions, clone H14R with an affinity of 220 pM (Jost and Plückthun, unpublished data) exhibited a similar, perhaps slightly weaker staining of HER2-amplified carcinomas when compared to the clone G3 (Figure 2e and f), whose affinity was determined as 90 pM.
HER2 Amplification Status Correlates Strongly with DARPin and Antibody
We analyzed 792 primary invasive carcinomas of the breast on tissue microarrays by HER2 fluorescent in situ hybridization, immunohistochemistry and DARPin-histochemistry using the clone G3 with direct detection of the stoichiometric biotinylation without further signal amplification (Table 1). Both antibody and DARPin correlated strongly with HER2 amplification status determined by fluorescent in situ hybridization as well as with each other (P<0.0001, each). Interestingly, we found the strongest correlation between DARPin and fluorescent in situ hybridization (0.87±0.02, Kendall's tau-beta).
DARPin-Histochemistry Results Increased Specificity Compared to Immunohistochemistry
In the present setup with only one biotin per DARPin, we found a significantly weaker staining on the tissue microarrays when comparing with the intensity of the immunohistochemistry staining, using the standard multimeric HRP coupled to the secondary antibody (P<0.0001) (Table 1, Figure 3). Most (97.8%) of the HER2 carcinomas found positive 1+ by immunohistochemistry were found to be negative by DARPin-histochemistry. Therefore, we conclude that a negative DARPin-histochemistry pattern (scored as 0) corresponds to a staining pattern of 0 or 1+ by immunohistochemistry and actually signifies an absence of amplification of HER2, as confirmed by fluorescent in situ hybridization. In accordance with this observation, we found in normal mammary glands no staining at all by DARPin-histochemistry (n=29), in contrast with the rather frequently encountered 1+ staining pattern by immunohistochemistry (n=8/33; data not shown).
Among the group of tumors scored as amplified by fluorescent in situ hybridization, we found in 97.4% cases either a 2+ or 3+ staining pattern by DARPin-histochemistry, whereas these tumors scored 3+ by immunohistochemistry. To better compare the HER2 amplification status by fluorescent in situ hybridization with HER2 expression measured by immunohistochemistry and DARPin-histochemistry, we therefore modified the scoring system (negative, equivocal and positive) for DARPin-histochemistry as follows: 0=negative, 1+=equivocal, and 2+ to 3+=positive.
Darpin G3 Detects Amplification Status of HER2 As Reliable As the FDA-Approved Antibody 4B5
Using the adapted scoring system for DARPin-histochemistry, we compared the amplification status determined by fluorescent in situ hybridization with the scored HER2 expression data determined by immunohistochemistry and DARPin-histochemistry (Table 2). Again, we found a strong correlation between the results obtained by DARPin-histochemistry, immunohistochemistry and amplification status measured by fluorescent in situ hybridization (P<0.0001, each). Among the HER2-amplified tumors, 86.2% were scored positive by immunohistochemistry, whereas 93.1% of these tumors were scored positive by DARPin-histochemistry. In the group of unamplified tumors, 99.4% were also scored negative by DARPin-histochemistry, in comparison with only 97.5% by immunohistochemistry.
We performed a receiver-operating characteristic analysis to check how reliably an equivocal or positive result by DARPin-histochemistry and immunohistochemistry could identify a positive amplification status (Figure 4), as this is important for decision on therapy. Both antibody and DARPin reliably identified HER2-amplification with similar sensitivities (98.9% for DARPin-histochemistry and 96.6% for immunohistochemistry). The specificity for the DARPin to identify a positive amplification status was significantly higher than that for the antibody (98.6% compared to 96.2%, P=0.0005, McNemar test).
Discussion
In this study, we report that a DARPin selected to specifically bind HER2 with picomolar affinity can reliably identify the amplification status of the HER2 proto-oncogene. DARPin-histochemistry proved to be as reliable and even displayed higher specificity when compared to an FDA-approved antibody for HER2 testing. In addition, we determined factors that will be important for developing and evolving such DARPin-based diagnostic tools.
Molecular Format and Affinity
When we compared the staining intensity of mono-, bi- and trivalent DARPins, where the monovalent one already had an affinity of 90 pM,27 we found that increasing avidity did not increase the intensity of staining (Figure 2e), even though we know that bivalent binding of these bivalent G3 DARPins can occur on tumor cells and that all epitopes are accessible.29 Although we cannot rigorously rule out steric effects at the binding site in the tissue samples, we note that at a concentration of 5 μg/ml (approximately 0.25–0.31 μM) the DARPin is already 200-fold above the KD of about 90 pM. Therefore, we conclude that an avidity effect does not provide greater binding, as HER2 is already saturated.
We also studied independent DARPins, G3, H6-3-B3 and H14R, for staining intensity and specificity on tissue microarrays, which all recognize epitopes in the extracellular domain IV of HER2. Although G3 and H6-3-B3 recognize overlapping epitopes,26, 27 H14R binds to a different epitope in domain IV of the ECD of HER2 (Jost and Plückthun, unpublished data) (Figure 2f). Of these, G3 gave the best staining pattern correlating with its high affinity. At least from this limited set of binders to the same target antigen, there was no evidence for a preferred epitope for diagnostics, independent of affinity. We also wish to emphasize that the fact that these DARPins recognize a conformational epitope does not constitute a limitation. Antigen retrieval by protease treatment rather than by heat denaturation was a simple measure to maximize the signal (Figure 2c).
The intensity of the staining of HER2-amplified carcinomas directly depended on affinity. G3 having an affinity of 90 pM yielded the most intense staining (Figure 2e), followed by H14R with an affinity of 220 pM (Figure 2f), whereas the correlation of staining intensity with affinity can nicely be seen in the point mutants of DARPin G3 with an affinity of ∼1, ∼10 and ∼270 nM (Figure 2e). Consistent with this observation, the DARPin H6-3-B3 with an affinity of 28 nM for HER2 failed to stain HER2-amplified tumors (Figure 2f and Supplementary Figure 2), but this could be overcome by raising the DARPin concentration in the detection solution. In conclusion, DARPins with affinities in the picomolar (and perhaps low-nanomolar) range are useful in diagnostics. Fortunately, with methods such as ribosome display such DARPins can be routinely obtained.17
Detection Method: Sensitivity and Specificity
Between the three detection methods compared here, we found that the detection by biotin-streptavidin, which is often the detection of choice on paraffin-embedded tissue sections, was also the most useful in our study. The highest specificity was achieved when the detection method was direct by the covalently bound biotin at the engineered AviTag with HRP-labeled streptavidin. Using the high-affinity DARPin G3, the detection of HER2 was already quantitative under the chosen conditions of 5 μg/ml (compare the staining in Figure 2b). We conclude that binding probably cannot be improved further and a further increase in signal intensity can only be achieved by using other means of detection. Future engineering efforts will thus have to be devoted to recruiting either more enzyme molecules to the bound DARPin, yet without sacrificing specificity, or resort to other detection methods altogether.30,31
Diagnostic Power of DARPin-Based Recognition HER2 Amplification in Patient Samples
Using an adapted scoring system for the DARPin-histochemistry, we show that in this study a similar sensitivity and higher specificity is reached to recognize a positive amplification status when compared to the FDA-approved antibody (Figures 3 and 4). Although the HER2 expression data, obtained either by DARPin or antibody, showed a strong correlation with the HER2 amplification status determined by fluorescent in situ hybridization, we found among the group of equivocals in fluorescent in situ hybridization a wide distribution of the protein data sets (see Table 2). Possibly, as a result of unselected material retrieval, some tumors exhibited already some degree of autolysis. Because this process has a stronger impact on the integrity of proteins than DNA, it might explain, why a larger number of tumors displayed an equivocal HER2 amplification status without subsequent evidence of increased protein expression.14
Conclusions
Our results strongly indicate that DARPins can be used in clinical diagnostics and provide a valuable alternative to antibodies. We found that affinity is a very important parameter, with picomolar binding required for maximal detection. Because DARPins with such properties can be quite routinely generated by ribosome display, are very robust and can be produced in very large quantities, DARPins specific for other antigens may turn out to be very useful for many clinical applications.
References
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007;25:118–145.
Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–792.
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 2005;353:1659–1672.
Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med 2006;354:809–820.
Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005;353:1673–1684.
Paik S, Kim C, Wolmark N . HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med 2008;358:1409–1411.
Owens MA, Horten BC, Da Silva MM . HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 2004;5:63–69.
Yaziji H, Goldstein LC, Barry TS, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA 2004;291:1972–1977.
Paik S, Bryant J, Tan-Chiu E, et al. Real-world performance of HER2 testing—National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst 2002;94:852–854.
Perez EA, Suman VJ, Davidson NE, et al. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol 2006;24:3032–3038.
Dal Lago L, Durbecq V, Desmedt C, et al. Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol Cancer Ther 2006;5:2572–2579.
Vera-Roman JM, Rubio-Martinez LA . Comparative assays for the HER-2/neu oncogene status in breast cancer. Arch Pathol Lab Med 2004;128:627–633.
Roche PC, Suman VJ, Jenkins RB, et al. Concordance between local and central laboratory HER2 testing in the breast intergroup trial N9831. J Natl Cancer Inst 2002;94:855–857.
Sauter G, Lee J, Bartlett JM, et al. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol 2009;27:1323–1333.
Sarkar CA, Dodevski I, Kenig M, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA 2008;105:14808–14813.
Steiner D, Forrer P, Plückthun A . Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol 2008;382:1211–1227.
Zahnd C, Amstutz P, Plückthun A . Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods 2007;4:269–279.
Binz HK, Stumpp MT, Forrer P, et al. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol 2003;332:489–503.
Forrer P, Binz HK, Stumpp MT, et al. Consensus design of repeat proteins. Chembiochem 2004;5:183–189.
Binz HK, Amstutz P, Plückthun A . Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol 2005;23:1257–1268.
Binz HK, Kohl A, Plückthun A, et al. Crystal structure of a consensus-designed ankyrin repeat protein: implications for stability. Proteins 2006;65:280–284.
Interlandi G, Wetzel SK, Settanni G, et al. Characterization and further stabilization of designed ankyrin repeat proteins by combining molecular dynamics simulations and experiments. J Mol Biol 2008;375:837–854.
Merz T, Wetzel SK, Firbank S, et al. Stabilizing ionic interactions in a full-consensus ankyrin repeat protein. J Mol Biol 2008;376:232–240.
Amstutz P, Koch H, Binz HK, et al. Rapid selection of specific MAP kinase-binders from designed ankyrin repeat protein libraries. Protein Eng Des Sel 2006;19:219–229.
Kawe M, Forrer P, Amstutz P, et al. Isolation of intracellular proteinase inhibitors derived from designed ankyrin repeat proteins by genetic screening. J Biol Chem 2006;281:40252–40263.
Zahnd C, Pecorari F, Straumann N, et al. Selection and characterization of Her2 binding-designed ankyrin repeat proteins. J Biol Chem 2006;281:35167–35175.
Zahnd C, Wyler E, Schwenk JM, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol 2007;369:1015–1028.
Theurillat JP, Zürrer-Hardi U, Varga Z, et al. NY-BR-1 protein expression in breast carcinoma: a mammary gland differentiation antigen as target for cancer immunotherapy. Cancer Immunol Immunother 2007;56:1723–1731.
Zahnd C, Kawe M, Stumpp MT, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer Res 2010;70:1595–1605.
Gullberg M, Jarvius J, Olsson C, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 2002;20:473–477.
Söderberg O, Leuchowius KJ, Gullberg M, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 2008;45:227–232.
Acknowledgements
The skillful technical assistance of Mrs Martina Storz is greatly appreciated. We thank Dr Annemarie Honegger for the preparation of the structure models shown in Figure 1. J-P Theurillat was supported by a grant from the Gertrud Hagmann-Stiftung für Malignomforschung.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Andreas Plückthun is shareholder of Molecular Partners AG that commercializes the DARPin technology.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Theurillat, JP., Dreier, B., Nagy-Davidescu, G. et al. Designed ankyrin repeat proteins: a novel tool for testing epidermal growth factor receptor 2 expression in breast cancer. Mod Pathol 23, 1289–1297 (2010). https://doi.org/10.1038/modpathol.2010.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.103
Keywords
This article is cited by
-
Studies of the oligomerisation mechanism of a cystatin-based engineered protein scaffold
Scientific Reports (2019)
-
Alternative reagents to antibodies in imaging applications
Biophysical Reviews (2017)