Abstract
The microphthalmia transcription factor/transcription factor E (TFE)-family translocation renal cell carcinomas bear specific translocations that result in overexpression of TFE3 or TFEB. TFE3 fusion gene product overexpression occurs as consequence of different translocations involving chromosome Xp11.2, whereas TFEB overexpression is the result of the specific translocation t(6;11)(p21;q12), which fuses the Alpha gene to TFEB. Both TFE3 and TFEB are closely related members of the microphthalmia transcription factor/TFE-family, which also includes TFEC and microphthalmia transcription factor. These transcription factors have overlapping transcriptional targets. Overexpression of microphthalmia transcription factor has been shown to mediate the expression of cathepsin-K in osteoclasts. We hypothesize that the overexpression of the related TFE3 fusion proteins and TFEB in translocation renal cell carcinomas may have the same effect. We studied cathepsin-K in 17 cytogenetically confirmed microphthalmia transcription factor/TFE-family translocation renal cell carcinomas. Seven cases showed a t(6;11)(p21;q12), ten cases showed translocations involving Xp11.2; five cases t(X;1)(p11;q21) resulting in a PRCC–TFE3 gene fusion; three cases t(X;1)(p11;p34) resulting in a PSF–TFE3 gene fusion, one t(X;17)(p11;q25) resulting in an ASPL–TFE3 gene fusion, and one t(X;3)(p11;q23) with an unknown TFE3 gene fusion. As control we analyzed cathepsin-K in 210 clear cell, 40 papillary, 25 chromophobe renal cell carcinomas and 30 oncocytomas. All seven TFEB translocation renal cell carcinomas were labeled for cathepsin-K. Among the cytogenetically confirmed TFE3 translocation renal cell carcinomas, 6 out of 10 were positive. None of the other renal neoplasms expressed cathepsin-K. We conclude the following: (1) cathepsin-K is consistently and strongly expressed in TFEB translocation renal cell carcinomas and in 6 of 10 TFE3 translocation renal cell carcinomas. (2) Cathepsin-K immunolabeling in both TFE3 and TFEB translocation renal cell carcinomas distinguishes these neoplasms from the more common adult renal cell carcinomas, and may be a specific marker of these neoplasms. (3) These results further support the concept that the overexpression of TFE3 or TFEB in these neoplasms activates the expression of genes normally regulated by microphthalmia transcription factor in other cell types.
Similar content being viewed by others
Main
The recently described microphthalmia transcription factor/transcription factor E (MiTF/TFE) family translocation renal cell carcinomas comprise the majority of pediatric renal cell carcinomas but also occur in adults. MiTF/TFE family translocation renal cell carcinomas are characterized by specific chromosome aberrations involving the genes transcription factor E3 (TFE3) and transcription factor EB (TFEB). The TFE3 transcription factor gene maps to chromosomal region Xp11.2, whereas the TFEB transcription factor gene maps to chromosome 6p21.1, 2 Several distinct gene fusions of TFE3 have been reported,1, 3, 4 whereas only the specific t(6;11)(p21;q12) seems to involve the locus of TFEB.2 These translocations result in the contribution of strong promoters that cause overexpression of the TFE3 fusion proteins or the native TFEB, such that they become detectable by immunohistochemical analysis.5, 6
Both TFE3 and TFEB are closely related members of the MiTF/TFE transcription factor family, which also includes TFEC and MiTF. All these transcription factors bind specific target DNA sequences both as homodimers or heterodimers, and have overlapping transcriptional targets.7, 8, 9, 10 Recently, DNA binding and transcriptional studies in osteoclasts have shown a consistent ability of MiTF to modulate the cathepsin-K promoter through three consensus elements, increasing mRNA and protein level of this papain-like cysteine protease.11 As the target DNA sequences of MiTF overlap with those of TFE3 and TFEB, we postulated that the overexpressed TFE3 fusion proteins or native TFEB in these renal cell carcinomas may have the same effect on the promoter of cathepsin-K. Therefore, in this study we have evaluated the expression of cathepsin-K in MiTF/TFE family translocation renal cell carcinomas and its potential usefulness as an immunohistochemical marker in their differential diagnosis from other renal cell carcinomas with which they may be confused.
Materials and methods
Tissue Samples
We collected 17 cases of MiTF/TFE translocation renal cell carcinomas (Table 1). All the cases had been earlier reported and cytogenetically confirmed.2, 3, 5, 12, 13, 14, 15, 16, 17 Seven cases harbored the translocation t(6;11)(p21;q12), which results in an Alpha-TFEB gene fusion and all showed confirmatory TFEB nuclear immunoreactivity as described earlier.5 Ten cases had translocations involving Xp11.2, and all ten of these renal cell carcinomas showed confirmatory TFE3 nuclear immunoreactivity as described earlier.6 Five cases showed the t(X;1)(p11.2;q21) resulting in the PRCC–TFE3 gene fusion; three cases showed the t(X;1)(p11.2;p34), resulting in the PSF–TFE3 fusion gene; one case showed the t(X;17)(p11.2;q25), resulting in the ASPL–TFE3 fusion gene and one case showed the novel t(X;3)(p11;q23).13For eight cases (cases 1, 2, 6–8, 10–12), five 1.4 mm diameter cores from the paraffin blocks of each case were used to create tissue microarrays for immunohistochemical analysis. These tissue microarrays also included six clear cell renal cell carcinomas and five papillary renal cell carcinomas. For the other nine cases (case 3–5, 9, 13–17), immunohistochemistry was performed in whole section slides from selected blocks. As control we collected 305 renal cell neoplasms from the files of the Renal Tumor Registry of the Department of Pathology of the University of Verona, including 210 clear cell renal cell carcinomas, 40 papillary renal cell carcinomas, 25 chromophobe renal cell carcinomas and 30 renal oncocytomas. Immunohistochemical labeling was also evaluated in the adjacent non-neoplastic renal tissues from the kidneys bearing the renal cell carcinomas.
Immunohistochemical Analysis
All tissue samples were formalin-fixed and paraffin embedded according to the standard methods. The sections from tissue blocks of MiTF/TFE family translocation carcinomas and control cases were immunolabeled with cathepsin-K using two different antibodies (clone CK4, Novocastra, Newcastle, UK and clone 3F9, Abcam, Cambridge, UK) and MiTF (clone D5, Neomarkers/LabVision, Fremont, CA, USA), using previously described methods.18 Heat-induced antigen retrieval was performed using a microwave oven and 0.01 mol/l of citrate buffer, pH 8.0, for 30 min. All samples were processed using a sensitive ‘Bond polymer Refine’ detection system in an automated Bond immunostainer (Vision-Biosystem, Menarini, Florence, Italy). Sections incubated without the primary antibody served as a negative control. Immunolabeling for cathepsin-K in the osteoclasts present in 10 specimens of remodeling bone was the positive control.
Results
Clinico-Pathological Findings
Clinico-pathological findings are summarized in Table 1. The tumors harboring translocations of chromosome Xp11 ranged from 3.1 to 13 cm in diameter and occurred in five women and five men (age range: 3–68 years; mean age: 19 years), whereas those harboring the t(6;11) occurred in five women and two men (age range: 6–54 years; mean age: 21 years).
Histological Findings
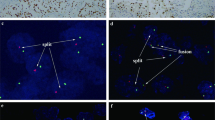
The Xp11 translocation carcinoma with novel break points, t(X;3)(p11;q23) (case 10), as reported earlier shared the same morphological features of the ASPL-TFE3 renal carcinomas (case 9), showing nested to papillary architecture composed of clear cells with voluminous cytoplasm and extensive psammomatous calcification.13 The PRCC-TFE3 renal carcinomas showed a more nested compact architecture. Psammoma bodies were rare or absent in these tumors. PSF-TFE3 renal carcinomas were composed of large polygonal clear cells in a mixed compact and papillary pattern. All 10 of these cases were immunoreactive for TFE3 protein. Two t(6;11) renal cell carcinomas were composed of polygonal elements, organized in nests and tubules, showing a sub-population of smaller cells with dark nuclei, frequently clustering around hyaline material. The other t(6;11) renal cell carcinomas had papillary architecture but maintained the dimorphic cytology (Figure 1a–d). All seven of these cases were immunoreactive for TFEB protein. MiTF was negative in all 17 neoplasms.
Immunohistochemical Findings
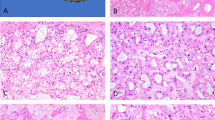
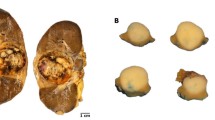
Immunohistochemical findings are summarized in Table 2. All seven of the renal cell carcinomas bearing the t(6;11) showed strong, diffuse cytoplasmic labeling for cathepsin-K with a granular pattern in 80–100% of the cells. In addition, dot-like cytoplasmic immunolabeling was observed in six of these cases. Among the cytogenetically confirmed Xp11 translocation renal cell carcinomas, 6 out of 10 were positive for cathepsin-K. All five renal cell carcinoma cases bearing the fusion product PRCC-TFE3 and one of the three renal cell carcinomas bearing the fusion product PSF-TFE3 were positive showing a granular pattern in 70–100% of the neoplastic cells (Figure 1e–h). None of the clear cell renal cell carcinomas, papillary renal cell carcinomas, chromophobe renal cell carcinomas or renal oncocytomas (Figure 2a–d) and none of the adjacent non-neoplastic renal tissue labeled for cathepsin-K (Figure 2e–h) or MiTF. The cytoplasm of occasional-activated macrophages within the neoplastic tissue of MiTF/TFE family translocation renal cell carcinomas or in the papillary cores of the papillary renal cell carcinomas were reactive for cathepsin-K, serving as an internal positive control. The immunohistochemical analysis with the two antibodies for cathepsin-K showed identical results. All the five tissue microarray cores from each case showed concordant immunohistochemical results (Figure 3).
All the usual renal cell tumors tested as control were negative for cathepsin-K. (a, e): Clear cell renal cell carcinoma, (b, f): Papillary renal cell carcinoma, note cathepsin-K immunolabeling the macrophages within the papillary cores. (c, g): Chromophobe renal cell carcinoma. (d, h): Renal oncocytoma.
All the five tissue microarrays cores from each case showed concordant immunohistochemical results. (a, d): Tissue microarrays cores from a papillary renal cell carcinoma and a Clear cell renal cell carcinoma negative for cathepsin-K. (b, c, e): MiTF/TFE family translocation renal cell carcinomas showing strong and diffuse immunolabeling for cathepsin-K (Cases 7, 2 and 11).
Discussion
In this study, we show that cathepsin-K is expressed in all seven t(6;11) renal cell carcinomas bearing the Alpha-TFEB gene fusion and in all five of the most common TFE3-positive renal cell carcinomas, those bearing the t(X;1)(p11.2;q21) and characterized by the fusion product PRCC-TFE3. Five of six TFE3-positive renal cell carcinomas harboring different translocations (three t(X;1)(p11.2;p34) renal cell carcinomas, one t(X;17)(p11.2;q23) and one t(X;3) renal cell carcinomas) did not label for cathepsin-K. Overall, 13 of 17 (76%) MiTF/TFE family translocation renal cell carcinomas were positive for cathepsin-K, whereas none of the 305 other renal cell neoplasms representing the most common renal cell neoplasms subtypes were not positive.
The rationale of our study was based on studies of osteoclast biology suggesting direct stimulation of cathepsin-K expression by MiTF.11 As MiTF is closely related to TFE3 and TFEB, having overlapping genomic targets, we postulated that in MiTF/TFE family translocation renal cell carcinomas cathepsin-K expression could be stimulated by these aberrantly expressed transcription factors. Different translocations involving chromosome Xp11.2 could result in different expression levels and structures of the TFE3 fusion products, potentially explaining why not all the TFE3-positive carcinomas express cathepsin-K. TFEB may be a stronger transcription factor in terms of driving cathepsin-K expression.
To date, immunohistochemistry for TFE3 and TFEB is the most reliable test able to distinguish MiTF/TFE family translocation renal cell carcinomas from formalin-fixed and paraffin-embedded archive tissue, but sometimes troubles using these antibodies have been reported.6, 19 Fluorescence in situ hybridization can represent an alternative method for detecting these translocations, as recently suggested, but the method has not been yet validated or standardized.16, 20 Our report shows that immunohistochemical analysis for cathepsin-K is a highly specific marker (though not completely sensitive) that can be helpful in the recognition of MiTF/TFE family translocation renal cell carcinomas from other renal cell carcinomas. On the basis of our results, a positive result strongly supports the diagnosis of translocation renal cell carcinomas, although a negative result does not exclude the diagnosis. Finally, our results support the concept that the expression profile of translocation renal cell carcinomas dramatically differs from that of the usual renal cell carcinomas. Further evidence for this concept comes from the not-infrequent expression of melanocytic markers HMB45 and Melan A, two targets normally activated by MiTF in melanocytes, in translocation renal cell carcinomas. Indeed, the global gene expression profile of translocation renal cell carcinomas more closely resembles alveolar soft part sarcoma than usual renal cell carcinomas.21 These data collectively suggest that the therapeutic targets against usual renal cell carcinomas may not be effective against translocation renal cell carcinomas due to the relevant differences occurring in the molecular pathogenesis of these carcinomas.
In conclusion (1) cathepsin-K appears to be consistently and strongly expressed in TFEB translocation carcinomas. (2) Cathepsin-K is expressed in 6 out of 10 TFE3 translocation carcinomas. It is possible that expression of cathepsin-K may be related to the specific TFE3 fusion harbored by the neoplasm. (3) Cathepsin-K immunoexpression in both TFE3 and TFEB translocation carcinomas distinguishes these neoplasms from the more common adult renal cell carcinomas. (4) These results suggest that the overexpression of TFE3 fusion proteins or native TFEB in these renal neoplasms activates expression of genes normally regulated by MiTF in other cell types.
References
Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol 2001;159:179–192.
Argani P, Hawkins A, Griffin CA, et al. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol 2001;158:2089–2096.
Argani P, Antonescu CR, Couturier J, et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am J Surg Pathol 2002;26:1553–1566.
Clark J, Lu YJ, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997;15:2233–2239.
Argani P, Lae M, Hutchinson B, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol 2005;29:230–240.
Argani P, Lal P, Hutchinson B, et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol 2003;27:750–761.
Fisher DE, Carr CS, Parent LA, et al. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev 1991;5:2342–2352.
Hemesath TJ, Steingrimsson E, McGill G, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994;8:2770–2780.
Kuiper RP, Schepens M, Thijssen J, et al. Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res 2004;32:2315–2322.
Zhao GQ, Zhao Q, Zhou X, et al. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol Cell Biol 1993;13:4505–4512.
Motyckova G, Weilbaecher KN, Horstmann M, et al. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci USA 2001;98:5798–5803.
Argani P, Lae M, Ballard ET, et al. Translocation carcinomas of the kidney after chemotherapy in childhood. J Clin Oncol 2006;24:1529–1534.
Argani P, Olgac S, Tickoo SK, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol 2007;31:1149–1160.
Onder AM, Teomete U, Argani P, et al. PRCC-TFE3 renal cell carcinoma in a boy with a history of contralateral mesoblastic nephroma. Pediatr Nephrol 2006;21:1471–1475.
Parast MM, Eudy G, Gow KW, et al. A unique case of renal carcinoma with Xp11.2 translocations/ TFE3 gene fusions in a 3-year-old child, with coexistent von Hippel-Lindau gene mutation. Pediatr Dev Pathol 2004;7:403–406.
Pecciarini L, Cangi MG, Lo Cunsolo C, et al. Characterization of t(6;11)(p21;q12) in a renal-cell carcinoma of an adult patient. Genes Chromosomes Cancer 2007;46:419–426.
Perot C, Boccon-Gibod L, Bouvier R, et al. Five new cases of juvenile renal cell carcinoma with translocations involving Xp11.2: a cytogenetic and morphologic study. Cancer Genet Cytogenet 2003;143:93–99.
Chilosi M, Poletti V, Zamo A, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol 2003;162:1495–1502.
Zhong M, Habermann J, Andraws N, et al. Xp11.2 translocation renal cell carcinoma (RCC) in adults - A TMA study of 120 RCC cases. Mod Pathol 2009;203A.
Mosquera JM, Dal Cin P, Mertz KD, et al. Validation of a TFE3 break-apart FISH assay in Xp11.2 translocation renal cell carcinomas. Mod Pathol 2008;172A.
Lae M, Argani P, Olshen A, et al. Global gene expression profiles of renal carcinomas with Xp11 translocations (TFE3 gene fusions) suggest a closer relationship to alveolar soft part sarcomas than to adult type renal cell carcinomas. Mod Pathol 2004;163A.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martignoni, G., Pea, M., Gobbo, S. et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol 22, 1016–1022 (2009). https://doi.org/10.1038/modpathol.2009.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.58
Keywords
This article is cited by
-
TSC loss is a clonal event in eosinophilic solid and cystic renal cell carcinoma: a multiregional tumor sampling study
Modern Pathology (2022)
-
Uterine PEComas: correlation between melanocytic marker expression and TSC alterations/TFE3 fusions
Modern Pathology (2022)
-
TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms
Virchows Archiv (2022)
-
TFEB rearranged renal cell carcinoma. A clinicopathologic and molecular study of 13 cases. Tumors harboring MALAT1-TFEB, ACTB-TFEB, and the novel NEAT1-TFEB translocations constantly express PDL1
Modern Pathology (2021)
-
TRIM63 is a sensitive and specific biomarker for MiT family aberration-associated renal cell carcinoma
Modern Pathology (2021)