Abstract
Recently, initial studies describing the use of multicolor fluorescence in situ hybridization (FISH) for classifying melanocytic skin lesions have been published demonstrating a high sensitivity and specificity in discriminating melanomas from nevi. However, the majority of these studies included neither histologically ambiguous lesions nor a clinical long-term follow up. This study was undertaken to validate a special multicolor FISH test in histologically ambiguous melanocytic skin lesions with known clinical long-term follow up. FISH was scored by three independent pathologists in a series of 22 melanocytic skin lesions, including 12 ambiguous cases using four probes targeting chromosome 6p25, centromere 6, 6q23, and 11q13. The FISH results were compared with array comparative genomic hybridization data and correlated to the clinical long-term follow up (mean: 65 months). Pair-wise comparison between the interpretations of the observers showed a moderate to substantial agreement (κ 0.47–0.61). Comparing the FISH results with the clinical behavior reached an overall sensitivity of 60% and a specificity of 50% (χ2=0.25; P=0.61) for later development of metastases. Comparison of array comparative genomic hybridization data with FISH analyses did not yield significant results but array comparative genomic hybridization data demonstrated that melanocytic skin lesions with the development of metastases showed significantly more chromosomal aberrations (P<0.01) compared with melanocytic skin lesions without the development of metastases. The FISH technique with its present composition of locus-specific probes for RREB1/MYB and CCND1 did not achieve a clinically useful sensitivity and specificity. However, a reassessment of the probes and better standardization of the method may lead to a valuable diagnostic tool.
Similar content being viewed by others
Main
Malignant melanoma is one of the most aggressive cancers that has an increasing incidence worldwide. Studies from Europe and the United States suggest a consistent and dramatic increase since the 1950s at about 3% per year.1, 2 More than 62 000 new cases of melanoma were predicted by the American Cancer Society to develop in the United States in 2008, with some 8400 deaths due to melanoma.3 Melanoma only contributes to about 4% of all skin cancer, but is responsible for 77% of deaths caused by skin cancer.4 Treatment continues to pose a substantial clinical challenge.
Melanocytic lesions exhibit a broad spectrum of biology ranging from definitive benign melanocytic nevi to highly aggressive melanoma. The diagnostic gold standard for the classification of melanocytic lesions and consideration of treatment options as well as disease prognosis still primarily relies on microscopic tissue morphology and depth of involvement. In most cases morphological criteria permit a definitive diagnosis of either benign or malignant cutaneous melanocytic lesions, but some lesions are notoriously difficult to diagnose by histology.5, 6 The Dutch melanoma working party reanalyzed a large cohort of 1069 patients with skin lesions originally classified by local pathologists to identify the most common diagnostic problems. It is interesting to note that in 8% of the cases the referring pathologist could not provide a confident diagnosis, in 14% the primary ‘malignant’ diagnosis had to be revised to ‘benign’ and in 17% the primary ‘benign’ diagnosis had to be revised to malignant.7
Recently, genetic approaches have been reported to be of significant help for classifiying melanocytic tumors. Comparative genomic hybridizations (CGHs) demonstrated that specific chromosomal aberrations in melanoma were distinct from chromosomal alterations in nevi.8 However, CGH is an expensive, time-consuming, non-morphology-based technique whose application in the current form is questionable for routine use.
A less expensive and easier screening method is fluorescence in situ hybridization (FISH). Using multicolor probes, this technology allows the detection of multiple genes at the same time. Initial studies analyzing the clinical applicability of such probes have been published, and demonstrated a very high sensitivity and specificity in distinguishing melanomas from nevi.9, 10, 11, 12 However, most of these studies did not examine ambiguous lesions or address clinical long-term outcomes. This study was undertaken to validate a special multicolor FISH kit in histologically ambiguous melanocytic lesions. The results were compared with the array CGH analysis and correlated to the clinical long-term follow up of the patients.
Materials and methods
Tumor Material and Clinical Data
A series of 22 melanocytic skin lesions that had been diagnosed between 1993 and 2008 was collected from the Department of Dermatology, University of Heidelberg, from the Dermatopathologische Gemeinschaftspraxis, Friedrichshafen and from the local pathologists. To get the case number presented in this study we screened files of melanoma patients, who visited the Department of Dermatology, University of Heidelberg for oncological treatment or aftercare within 12 months (July 2007–June 2008).
All patients with ambiguous melanocytic lesions, with complete clinical follow up, and available tumor material were included in this retrospective study.
All lesions had been resected owing to clinical suspicion of malignancy. After complete excision, standard histological examination was carried out on formalin-fixed, paraffin-embedded tissue sections using hematoxylin and eosin (H&E) staining. Melanoma tumor staging was determined according to the latest TNM-staging system.13 The clinical and histological features of the patients are summarized in Table 1. The mean observation period of all patients was 65 months (range 10–156) and 71 months for the ambiguous cases. A melanocytic skin lesion was considered to be of malignant clinical behavior if lymphatic, hematologic, in transit, or satellite metastases occurred. Melanocytic skin lesions without the detection of metastases were classified as showing a benign clinical behavior. A local relapse was not included as a sign of malignant behavior. Depending on the stage of disease, the clinical examinations were carried out on the basis of standard recommended melanoma specific follow-up care.14
Accordingly, the 22 melanocytic skin lesions were categorized into five groups:
-
1)
Histologically benign nevi with benign clinical behavior (n=3).
-
2)
Histologically ambiguous melanocytic skin lesions with benign clinical behavior (n=7).
-
3)
Histologically ambiguous melanocytic skin lesions with malignant clinical behavior (n=5).
-
4)
Histologically definitive malignant melanomas with benign clinical behavior (n=4).
-
5)
Histologically definitive malignant melanomas with malignant clinical behavior (n=3).
The patient group consisted of 9 males and 13 females with a median age of 49 years (range 17–68). The median tumor thickness in all melanocytic skin lesions was 1.6 mm (range 0.4–2.8) and most lesions reached Clark level IV. Indeterminate histological diagnosis most often occurred between Spitz nevus vs Spitzoid melanoma, followed by melanoma with regression vs atypical nevus.
FISH
Probes targeting four loci were used for FISH evaluation. These included ras responsive element binding protein 1 (RREB1) on 6p25 (Vysis LSI RREB1-Spectrum red), V-myb myeloblastosis viral oncogene homolog (MYB) on 6q23 (Vysis LSI MYB-Spectrum gold), cyclin D1 (CCND1) on 11q13 (Vysis LSI CCND1-Spectrum green) and centromeric enumeration probe control for chromosome 6 (Vysis LSI CEP6-Spectrum aqua) from Abbott Molecular, Des Plaines, IL, USA.
Consecutive 4 μm thick sections were obtained from the same blocks as those used for H&E staining. Sections were cut and mounted on SuperFrost+/+ slides. Deparaffinising, pre-treatment, and protease digestion procedures were carried out according to the Vysis manual. After hybridization of the probe mixture at 37°C for 16–24 h, slides were washed in 2 × SSC/0.3% NP-40 stringency buffer at 72°C for 2 min. Subsequently, slides were air dried and counterstained with DAPI. FISH evaluation was carried out according to the Vysis manufacturer's protocol. For the evaluation of gene amplification, 30 randomly selected, non-overlapping nuclei of invasive tumor cells in three separate, and distinct tumor areas (10 nuclei in each area) were scored for the number of fluorescent signals using the Olympus BX 50 Fluorescence/Phase Microscope (TG) and the Zeiss Axioplan-2 Microscope (HK, GP). A case was considered as malignant melanoma if one of the following from the manufacturer's established criteria was fulfilled:
-
a)
Average number of CCND1 signals per nucleus ≥2.5
-
b)
Average number of MYB signals per nucleus ≥2.5
-
c)
Percentage loss of MYB in relation to CEP6 ≥31%
-
d)
Percentage of atypical nuclei for RREB1 ≥63%
The FISH evaluation was done by three independent pathologists, all long-time experienced in FISH diagnostics (HK, GP in Friedrichshafen, and TG in Heidelberg), blinded against primary histological diagnosis, clinical follow-up, and previously gained FISH results. Every pathologist evaluated each LSI probe (RREB1, CCND1, MYB, and centromere 6).
To correlate the FISH results with histology and clinical behavior, a consensus score was built. A lesion was considered as FISH positive when at least two of the three observers diagnosed a positive FISH result.
Array CGH
In 16 cases, a genome wide analysis of genomic aberrations was carried out using a high-resolution array CGH (array CGH) technique as described previously.15 To obtain a high purity of tumor DNA, only areas with a tumor cell infiltration of over 90% were microdissected for DNA isolation using the QIAamp DNA formalin-fixed, paraffin-embedded tissue kit (Qiagen, Hilden, Germany). Samples were labeled with a bioprime array CGH genomic labeling kit according to the manufacturer's instructions (Invitrogen, Carlsberg, CA, USA). Briefly, 500 ng test DNA and reference DNA were differentially labelled with dCTP-Cy5 and dCTP-Cy3, respectively (GE Healthcare, Piscataway, NJ, USA). Genome-wide analysis of DNA copy number changes was carried out using an oligonucleotide array containing 44 000 probes with a spatial resolution of 35 kb according to the manufacturer's protocol version 6.0 (Agilent, Santa Clara, CA, USA). Slides were scanned with Agilent's microarray scanner G2505B and analyzed using Agilent CGH Analytics software 4.0.76 (statistical algorithm: ADM-2; sensitivity threshold: 6.0; consecutive clone filter: 20).
Statistics
Data was entered into Microsoft EXCEL 2003; statistical analysis was done with SAS version 9.1WIN and with SPSS version 16.0. Concordance between FISH data, array CGH data, histology, tumor thickness, and clinical follow up was determined by using the Fisher's Exact Test, McNemar's Test, odds ratio, and Student's t-test. Unvaried variance analyses and linear regression were calculated. To evaluate inter-observer variability the κ statistics were calculated with Cohen's κ. A κ value of 0.00–0.20 indicates slight agreement; 0.21–0.40 fair agreement; 0.41–0.60 moderate agreement; 0.61–0.80 substantial agreement; and 0.81–1.00 almost perfect agreement.
Results
Histological Diagnosis
In all, 12 out of the 22 investigated samples could not be diagnosed as malignant or benign because of ambiguous histological features. Seven cases were definitely diagnosed as malignant melanomas and three cases were determined to be benign nevi (further details of the histological diagnoses are provided in Table 1).
Failure Rate and FISH Inter-Observer Variability
In 16 of the 22 investigated cases, FISH data could be evaluated by all the three pathologists. Because of insufficient signals, four cases had to be excluded and owing to signal fade two samples were evaluated by only one investigator. Overall agreement among the three pathologists was achieved in 11 of 16 cases (68%): 5 positive FISH results and 6 negative FISH results. Pair-wise comparison between observer interpretations showed a moderate to substantial agreement (Table 2). Observers HK and TG were in accordance in 13 of 16 cases (81%), HK and GP were in accordance in 13 of 16 cases (81%), and TG and GP were in accordance in 12 of 16 cases (75%).
FISH Results and Correlation with Clinical Behavior
Multitarget FISH gave a positive consensus result (≥1 FISH criterion positive) in 7 of 16 melanocytic skin lesions. An example is given in Figure 1. In detail, 5 melanocytic skin lesions were positive for MYB-loss, 4 melanocytic skin lesions were positive for RREB1 amplification, 2 melanocytic skin lesions were positive for CCND1 and 1 melanocytic skin lesion was positive for MYB amplification. In total, 1 melanocytic skin lesion was positive for 3 criteria (CCND1, MYB, and MYB-loss), 2 melanocytic skin lesions were positive for 2 criteria, and 5 melanocytic skin lesions were positive for 1 criterion. Two of the seven ambiguous melanocytic skin lesions without metastases were FISH positive (four without signals, one FISH negative) and three of the five ambiguous melanocytic skin lesions with metastases were FISH positive. The four histologically definitive melanomas without metastases yielded two positive FISH results, and notably only one of the definitive three melanomas with metastases yielded a positive FISH result. All three benign nevi were FISH negative (for details see Table 1).
Comparing the FISH results with the clinical behavior of the melanocytic skin lesions gave an overall sensitivity of 60% and a specificity of 50% (χ2=0.25, P=0.61). Neither the univariate analysis of variance nor the linear regression analysis yielded a significant result for the parameters of histological diagnosis, Breslow tumor thickness, Clark-level, or FISH status (data not shown).
Array CGH Results and Correlation with Clinical Behavior
Array CGH could be carried out in 8 of the 16 samples that had been investigated by all the three observers. The other 8 samples failed because of insufficient remaining tumor material or degradation of DNA. The number of chromosomal aberrations per tumor ranged from 6 to 65 (mean 10.5).
Depending on the clinical follow up, but not on the previous histological diagnosis, two groups of melanocytic skin lesions were generated:
-
1)
Melanocytic skin lesions with malignant clinical behavior (n=3); and
-
2)
Melanocytic skin lesions with benign clinical behavior (n=5).
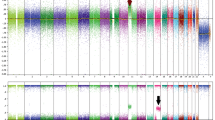
A striking increase of chromosomal aberrations (P<0.01) could be detected within the group of melanocytic skin lesions with malignant clinical behavior compared with the group of melanocytic skin lesions with benign clinical behavior despite the small number of cases per group (Figure 2).
It is interesting to note that all the three melanocytic skin lesions with malignant clinical behavior showed gains of chromosome 7p22, 9q34, 11p15, 11q13, 14q32, 16q13, 17q25, 19p13, and 20q13.
Array CGH Results and Comparison with FISH Data
Comparison of array CGH data and FISH analysis yielded incongruent data. Chromosomal aberrations, which had been detected with FISH at the according loci, were not reproducible with array CGH (Table 1). In contrast, loci that did not show chromosomal aberrations with FISH, yielded positive results with array CGH. One histologically definitive malignant melanoma that had yielded negative results with FISH analysis showed unaffected 6p25, 6q23, and 11q13 loci in the array CGH, thereby explaining the negative FISH result. However, this case displayed several chromosomal aberrations in other loci.
Discussion
Recently, modified molecular targeting allowed chromosomal analyses of routinely formalin-fixed, paraffin-embedded melanoma tissue for the first time. Initially, studies investigating the loss of heterozygosity16, 17 and later CGH studies8, 18, 19 detected chromosomal imbalances in primary cutaneous melanomas. The copy number changes in melanomas depend on body site and level of sun exposure.20 The most frequently gained regions in melanomas are found in 6p (37%), 1q (33%), 7p (32%), 7q (32%), 8q (25%), 17q (24%), and 20q (22%), whereas the most frequent losses are seen in chromosome 9p (64%), 9q (36%), 10q (36%), 10p (30%), 6q (26%), and 11q (21%).8 Melanocytic nevi show no chromosomal aberrations except for the Spitz nevus, which displays a gain of 11p in 20%.19 Based on these studies, a probe combination for LSI RREB1/LSI MYB/LSI CCND1/CEP 6 has been developed for detection of amplifications of 6p25 (RREB1), 6q23 (MYB), 11q13 (CCND1), and loss of 6q23 (MYB).
Before this method can be accepted as a routine diagnostic tool for critical melanocytic lesions, technical and clinical validation is critical.
To expand on the clinical validation of these probes already reported, we retrospectively investigated a series of melanocytic lesions with initially ambiguous histological results. The FISH results were further compared with the clinical long-term follow up (mean 71 months), thereby either encouraging or challenging the primary diagnosis. Histologically definitive cases with follow up were included as positive and negative controls. Our analysis of primary melanocytic lesions using the Vysis LSI FISH detection system compared with clinical outcomes yielded incongruent results. It was not possible to reach a clinical relevant sensitivity or specificity using this probe combination.
It is important to note that there were histologically definitive malignant melanomas with the consequent development of metastases during the clinical long-term follow up that yielded negative results with the FISH probes. As expected, none of the histologically definitive benign nevi were FISH positive. Our results seem contradictory to recent studies reporting high sensitivity and specificity of the four FISH probes.9, 10, 12 However, these studies did not include histologically ambiguous lesions, and the results of the FISH analysis had been compared with the primary histological diagnosis but not to the clinical long-term follow up, considered the diagnostic gold standard. That said, the development of metastases during the long-term follow up is not conclusive of the malignant potential of the primary melanocytic lesion, and may instead arise from a separate unknown primary elsewhere in the body. Conversely, the absence of metastases does not conclusively reflect the benignity of the primary lesion despite several years of clinical follow up. Development of melanoma metastases has been reported to occur even after 15 years of tumor-free status.21
However, one study did attempt to overcome these limitations by analyzing a large series of different melanocytic lesions including histologically ambiguous cases, FISH analysis, and clinical follow-up data.11 In this study the four color FISH test correctly classified melanoma with 87% sensitivity and 95% specificity. The test also correctly identified as melanoma 6 of 6 ambiguous cases that later metastasized. However, consistent with our findings of non-specificity, there were histologically ambiguous lesions with benign clinical behavior but positive FISH results. The authors’ explanation of these inconsistent findings is either a cure of melanoma by the removal of the primary tumor or an insufficiently long follow up for the detection of metastasis. A limitation of this study as well as our own investigation was the small number of cases with an ambiguous initial diagnosis correlated with a long-term follow up. But this constraint is mainly caused by the difficult access to this rare case collective.
Another limitation in the recently reported studies is that none of them demonstrated an inter-observer agreement. In our study, three pathologists, all experienced in FISH diagnostics independently examined all cases. Subsequently, a consensus score was generated to keep possible interpretation discrepancies as low as possible. The inter-observer variability showed a moderate to substantial agreement, which is a contenting result but could be upgraded by a better standardization of the evaluation of the FISH test.
To exclude the possibility the FISH probes miss other chromosomal aberrations, and as a control of the FISH results, a subgroup of cases was further evaluated with array CGH. Unfortunately, array CGH and FISH analysis yielded incongruent data. Chromosomal aberrations, which had been detected with FISH at the according loci, were not reproducible with array CGH. Conversely, the loci that did not show chromosomal aberrations with FISH yielded positive results with array CGH. A reason for the discrepancy between the FISH and array CGH data in our study may be the limited resolution of the array CGH. This method may detect chromosomal gains and losses from a 700 kb threshold leading to a possible oversight of aberrations in solitary genes. Furthermore, copy number changes must be present in a substantial proportion of cells (at least 25%) to be identifiable. This, again, is not a prerequisite for FISH analysis. Another reason for the discrepancy of the FISH and array CGH data may be the clonal heterogeneity that has been reported in melanoma,22 thereby exhibiting differing features on different sections of the tumor.
Interestingly, our histologic definitive melanoma yielded negative results with the three FISH probes. Consistent with this finding, array CGH showed unaffected loci 6p25, 6q23, and 11q13, thereby explaining the negative FISH results. It is interesting to note that this case displayed several chromosomal aberrations in other loci. As a side note, melanocytic skin lesions with metastases showed considerably more chromosomal aberrations with array CGH than melanocytic skin lesions without metastases.
In our opinion, the prediction of metastatic behavior in malignant melanoma based on only three chromosomal loci seems unlikely. This assumption is further emphasized in a study of Ramaswamy et al23 demonstrating by means of gene expression arrays that no less than a gene signature of 17 genes allows a reliable prediction of metastatic behavior of a primary tumor.
In summary, in our study the FISH probes did not detect all of the relevant chromosomal changes necessary for a clinically useful diagnostic aid to distinguish benign from malignant melanocytic lesions. A modification of the composition of the probes may overcome these limitations and lead to a superior diagnostic tool.
References
Geller AC, Miller DR, Annas GD, et al. Melanoma incidence and mortality among US whites, 1969–1999. JAMA 2002;288:1719–1720.
Stang A, Pukkala E, Sankila R, et al. Time trend analysis of the skin melanoma incidence of Finland from 1953 through 2003 including 16 414 cases. Int J Cancer 2006;119:380–384.
American Cancer Society. http://www.cancer.org/docroot/cri/content/cri_2_4_1x_what_are_the_key_statistics_for_melanoma_50.asp.
Abdulla FR, Feldman SR, Williford PM, et al. Tanning and skin cancer. Pediatr Dermatol 2005;22:501–512.
Farmer ER, Gonin R, Hanna MP . Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol 1996;27:528–531.
Spatz A, Ruiter D, Hardmeier T, et al. Melanoma in childhood: an EORTC-MCG multicenter study on the clinico-pathological aspects. Int J Cancer 1996;68:317–324.
Veenhuizen KC, De Wit PE, Mooi WJ, et al. Quality assessment by expert opinion in melanoma pathology: experience of the pathology panel of the Dutch Melanoma Working Party. J Pathol 1997;182:266–272.
Bastian BC, Olshen AB, LeBoit PE, et al. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol 2003;163:1765–1770.
Morey AL, Murali R, McCarthy SW, et al. Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology 2009;41:383–387.
Newman MD, Lertsburapa T, Mirzabeigi M, et al. Fluorescence in situ hybridization as a tool for microstaging in malignant melanoma. Mod Pathol 2009;22:989–995.
Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol 2009;33:1146–1156.
Newman MD, Mirzabeigi M, Gerami P . Chromosomal copy number changes supporting the classification of lentiginous junctional melanoma of the elderly as a subtype of melanoma. Mod Pathol 2009;22:1258–1262.
American Joint Committee on Cancer. http://www.cancerstaging.org/products/ajccproducts.html.
Garbe C, Schadendorf D, Stolz W, et al. Short German guidelines: malignant melanoma. J Dtsch Dermatol Ges 2008;6 (Suppl 1):S9–S14.
Geigl JB, Obenauf AC, Waldispuehl-Geigl J, et al. Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays. Nucleic Acids Res 2009;37:e105.
Isshiki K, Elder DE, Guerry D, et al. Chromosome 10 allelic loss in malignant melanoma. Genes Chromosomes Cancer 1993;8:178–184.
Millikin D, Meese E, Vogelstein B, et al. Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res 1991;51:5449–5453.
Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 1998;58:2170–2175.
Bastian BC, Wesselmann U, Pinkel D, et al. Molecular cytogenetic analysis of Spitz nevi shows definitive differences to melanoma. J Invest Dermatol 1999;113:1065–1069.
Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135–2147.
Tsao H, Cosimi AB, Sober AJ . Ultra-late recurrence (15 years or longer) of cutaneous melanoma. Cancer 1997;79:2361–2370.
Bogdan I, Xin H, Burg G, et al. Heterogeneity of allelic deletions within melanoma metastases. Melanoma Res 2001;11:349–354.
Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet 2003;33:49–54.
Acknowledgements
We thank Christine DeWitt (National Institutes of Health) for critical reading of the paper. The authors thank Abbott for generously providing the Melanoma FISH probe.
Trademarks: The multicolor FISH probe combination consists of 4 specific Locus specific identifiers (LSI) for RREB1, MYB, CCND1, CEP 6 and is a registered trademark of Abbott Molecular Incorporation.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Disclosure/conflict of interest
TG, MRB, and HK received honoraria from Abbott Molecular Labs. All authors state that there are no relevant conflicts of interest or funding sources to declare.
Rights and permissions
About this article
Cite this article
Gaiser, T., Kutzner, H., Palmedo, G. et al. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol 23, 413–419 (2010). https://doi.org/10.1038/modpathol.2009.177
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.177
Keywords
This article is cited by
-
Four-color fluorescence in-situ hybridization is useful to assist to distinguish early stage acral and cutaneous melanomas from dysplastic junctional or compound nevus
Diagnostic Pathology (2020)
-
Through the looking glass and what you find there: making sense of comparative genomic hybridization and fluorescence in situ hybridization for melanoma diagnosis
Modern Pathology (2020)
-
Comparison of melanoma gene expression score with histopathology, fluorescence in situ hybridization, and SNP array for the classification of melanocytic neoplasms
Modern Pathology (2018)