Abstract
MicroRNAs (miRNAs) are involved in cell proliferation, differentiation, and apoptosis and can function as tumor suppressor genes or oncogenes. The role of miRNAs in neuroendocrine tumors such as ileal carcinoids is largely unknown. We examined the differential expression of 95 miRNAs by RT–PCR using the QuantiMir System in eight matching primary and metastatic carcinoid tumors from the ileum. All miRNAs chosen for the QuantiMir System array were based on their potential functions related to cancer biology, cell development, and apoptosis. The expression of miRNAs for the samples was normalized to miRNA-197, and the matching primary and metastatic tumors were compared. There was downregulation of miRNA-133a, -145, -146, -222, and -10b in all samples between the primary and matching metastatic tumors and upregulation of miRNA-183, -488, and -19a+b in six of eight metastatic carcinoids compared to the primary tumors. miRNA-133a was further analyzed by TaqMan real-time RT–PCR and northern hybridization using six additional matching primary and metastatic samples, which supported the PCR array findings. There were significant differences in miRNA-133a expression with downregulation in the metastasis compared to the primary in the eight original cases (P<0.009) and in the six additional cases used for validation (P<0.014). Laser capture microdissection and real-time RT–PCR analysis using normal ileum found miRNA-133a expression in normal enterochromaffin cells. In situ hybridization in normal ileum showed that some of the mucosal endocrine cells expressed miRNA-133a. Both primary and metastatic ileal carcinoid tumors expressed miRNA-133a by in situ hybridization. These results provide information about novel marker miRNAs that may be used as biomarkers and/or therapeutic targets in intestinal carcinoid tumors.
Similar content being viewed by others
Main
MicroRNAs (miRNAs) are a family of 21–25 nucleotide, non-coding small RNAs that function as gene regulators.1, 2, 3, 4, 5, 6, 7 MiRNAs are usually excised from 60–110 nucleotide fold back RNA precursor molecules. They are involved in critical biologic processes including development, differentiation, apoptosis, and proliferation.1, 6, 7 MiRNAs regulate posttranscriptional gene silencing by inducing mRNA degradation or by binding to the 3′-untranslational region of target RNAs leading to repression of the translational process.1, 6, 7 Mature miRNAs are processed from stem-100 bp precursor molecules, which are transcribed as part of longer primary transcripts. The primary miRNAs are processed in the nucleus by the RNase Drosha, and then the precursor miRNAs are exported to the cytoplasm and further processed by the RNase Dicer.1, 6, 7 MiRNAs are usually deregulated in cancers.1, 6, 7
Gastroenteropancreatic neuroendocrine tumors, which include ileal carcinoids, are rare neoplasm, which secrete peptides and neuroamines, that can cause distinct clinical syndromes including the carcinoid syndrome.8, 9, 10, 11 The incidence of carcinoid tumors has been increasing i8 and recent studies have begun to elucidate the role of growth factors, growth factor receptors, and other molecules in the development of these tumors; however, the pathogenesis remains unknown.8, 9, 10, 11, 12, 13, 14, 15 Patients with advanced disease often present with metastatic disease to the liver and other sites. Many of these patients have unresectable disease and only a limited number of drugs are available for treatment of these patients.13, 16, 17
MiRNAs have not been previously examined in carcinoid tumors. We analyzed the type of miRNAs expressed in normal enterochromaffin cells from the terminal ileum and in primary midgut carcinoid and in metastatic ileal carcinoid tumors to determine whether specific miRNAs would have a unique profile in these neuroendocrine tumors and whether there were changes in the levels of miRNA expression in primary and metastatic carcinoids and to possibly identify important molecular tests for further functional investigations to develop new diagnostic and therapeutic targets for midgut carcinoid tumors.
Materials and methods
Tissues and miRNA Extraction
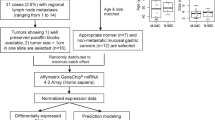
Tissues analyzed included eight fresh frozen matching primary and metastatic human ileal carcinoid tumors containing 70–90% tumor. Another six matching pairs of primary and metastatic ileal carcinoids were also used for validation studies. Approximately 100 mg of each tumor was disrupted using a mechanical homogenizer. Total RNA containing the small RNA fraction was then extracted by acid-pheno1:chloroform using the mirVana miRNA Isolation kit (Ambion Inc., Austin, TX, USA) or with Trizol (Invitrogen, Carlsbad, CA, USA). RNA was purified by glass-fiber filters included in the kit, quantified by spectrophotometric absorbance at A260 and stored at −70°C. RNA quality was assessed using the Small RNA kit (Agilent) on the Agilent 2100 Bioanalyzer. IRB approval was obtained for the study.
Real-Time RT–PCR Array
One microgram total RNA from each carcinoid tumor sample was reverse transcribed using the QuantiMir kit (System Biosciences (SMI), Mountain View, CA, USA) in a total volume of 20 μl.
The cDNA sample was analyzed by real-time PCR using the Cancer MicroRNA qPCR Array kit (SBI), which provides 95 miRNA-specific and U6-specific forward primers in a 96-well plate format and the 3′ universal reverse primer. SYBR Greener Real-Time PCR Super Mix (Invitrogen, La Jolla, CA, USA) was added to a final concentration of 1 × . Each cDNA sample was run in triplicate on a 384-well optical plate in a total volume of 15 μl per well. qPCR was performed on the 9700HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the following profile: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. A dissociation step was performed after the qPCR amplification for melting curve analysis.
Calculation of Relative Expression Level
Threshold cycle (Ct) values were defined as the cycle number at which the fluorescence exceeded a fixed threshold value above an automatically set baseline. MiR-197 was selected as the endogenous control because of Ct values that vary less than 1.0 cycle between matching primary and metastatic samples. Ct values of each target miRNA transcript were normalized against the Ct value of miR-197. Relative change of miRNA expression in metastatic compared to its primary carcinoid tumor was then calculated using the ΔΔCt method. Three replicates for each experimental sample were performed and averaged together. Ten individual miRNA targets did not amplify or give a clean melting curve and were excluded. Results were expressed as the mean±s.e.m. Melting curve analysis of the qPCR products verified product specificity.
Real-Time Rt–PCR TaqMan miRNA Expression Assay
MiRNA-133a was selected from the PCR array for further analysis. Mature miRNA expression levels were measured using the TaqMan MicroRNA Assay kit (Applied Biosystems). Total RNA of 10 ng from matching primary and metastatic carcinoids from the eight samples analyzed earlier and an additional six material samples were reverse transcribed in a total volume of 15 μl using the TaqMan MicroRNA Reverse Transcription kit according to the manufacturer's instruction; 1.33 μl of each RT reaction was amplified by PCR in 20 μl total containing 10 μl of the TaqMan 2 × Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). PCR was performed on the 7900HT Fast Real-Time PCR System with an initial incubation at 95°C for 15 s and 60°C for 60 s. Each PCR reaction was performed in triplicate a minimum of three times. Agarose gel electrophoresis and densitometric analyses were performed. Expression levels were normalized with a small nucleolar RNA, RNU48, which was co-amplified as the endogenous control.
Northern Blotting
The northern blot assay was performed using the MiRNA Northern Blot Assay kit (Signosis) according to the manufacturer's protocol. In brief, following a 30-min pre-run, 20 μg denatured total RNA from each matching tumor case was resolved by electrophoresis in a 60% TBE-Urea Ready Gel (Bio-Rad). After the transfer, the membrane was UV cross-linked and baked at 42°C in the hybridization buffer provided by the kit; 10 μl of the RNU48 DIG-labeled probe and 10 μl of hsa-miR-133a biotin-labeled probe were added and incubated overnight at 42°C. The blots were washed for 30 min then blocked for 30 min in blocking buffer. Streptavidin–horse radish peroxidase conjugate of 20 μl was added to the blocking buffer for 45 min. The blots were washed three times with detection washing buffer. An equal mixture of Substrate A and Substrate B was then applied to the blot for 5 min. The excess liquid was removed, and the blots exposed to Kodak radiography film and developed.
Laser Capture Microdissection of Normal Enterochromaffin Cells of Ileal Mucosa
Laser capture microdissection was combined with immunocytochemical staining using specific antibodies to recognize endocrine cells associated with the crypts. Normal ileal mucosa tissue blocks were cut in 10 μm (cryostat) sections and adhered on uncharged glass slides and single cells were isolated as described earlier.18, 19 Briefly, the slides were incubated with anti-chromogranin A antibody (1/100 Dako, Santa Barbara, CA, USA), washed, and incubated with a secondary horse radish peroxidase-conjugated antibody, washed, and counterstained with hematoxylin. All steps were performed at the minimum time required and always in the presence of RNAse inhibitor. We have verified in the past that our anti-chromogranin A antibody can be combined with laser capture microdissection.19 Enterochromaffin cells are located in the crypts within the ileum, and we estimate that one endocrine cell and 3–4 non-endocrine cells were captured with each laser treatment, ie, approximately 25% of the captured cells are endocrine and the majority of them are enterochromaffin cells. Immunostained cells were isolated using a PixCell II laser-capture microdissection system (Arcturus Bioscience, Mount View, CA, USA).
In Situ Hybridization
In situ hybridization to detect specific miRNAs was performed using normal ileum (n=5), frozen sections of the original eight matched cases used for miRNA RT–PCR array and qRT–PCR screening and an additional six cases of matched primary and metastatic tumors. In addition, 30 formalin-fixed paraffin-embedded tissue cases of matching primary and metastatic tumors to the liver were also analyzed. Probes for in situ hybridization were purchased from Exiqon (Denmark and Woburn, MA, USA) and consisted of locked nucleic acid oligonucleotide probes labeled at both the 5′ and 3′ ends with digoxigenin. In situ hybridization was performed as reported earlier from our laboratory20 with modifications for the locked nucleic acid probes.21, 22, 23 Frozen tissues were fixed in 4% paraformaldehyde and stored at −70°C until used.
Briefly, in situ hybridization was performed by microwave pretreatment in a 700 W oven in 10 mM citric acid pH 6.0 for 5 min (frozen tissues) or 12 min (paraffin-embedded tissues) followed by cooling at room temperature for 20 min. Tissues were digested in 25 μg/ml proteinase K at 37°C for 5 min (frozen tissues) or 10 min (formalin-fixed paraffin-embedded tissue) solution. Hybridization was performed at 60°C overnight in a humidified chamber using 100 nM for frozen tissues and 200 nM of probe for formalin-fixed paraffin-embedded tissues. Slides were washed in 2 × sodium chloride–sodium citrate twice for 3 min at 55°C and in 0.5 × 55°C twice for 10 min.
Slides were rinsed in Tris-buffered saline, pH7.5 (buffer A) for 3 min and treated with 300 μl of anti-digoxigenin-alkaline phosphatase antibodies at a 1/200 dilution for 3 h followed by washes in buffer A with 1% normal swine serum and 0.3% Triton X-100. After washes in buffer C, pH 9.5, the slides were reacted with nitroblue-tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate, pH9.5 in buffer C and developed between 1 and 4 h and counterstained with 0.1% Nuclear Fast Red for 3 min.
Controls consisted of substituting a scrambled probe (Exiqon) at the same concentration as the hybridization probe. As a second control, slides were treated with RNAse A before hybridization. Both the scramble probe and RNAse pretreatment resulted in no hybridization signal.
Slides were scored as 0, negative, 1+ weak staining, 2+ moderate staining, and 3+ strong staining.
Statistical Analysis
Gene Spring GX 10.0 software was used to identify miRNAs with a metastatic to primary ratio less than or greater than 1 (upregulated or downregulated in metastatic tumors) in all eight samples. An average of the ratio across all eight samples was calculated and miRNAs were ranked in order of increasing average.
Densitometric analysis after real-time RT–PCR and gel electrophoresis was expressed in mean ±s.e. of the mean and analyzed with the Student's t-test. P<0.05 was considered statistically significant.
Results
Clinicopathologic Features
The eight cases consisted of six females and two males. Metastatic tumor was present in the liver in seven cases and in a mesenteric lymph node in 1 case (Table 1). The six additional case studies included six primary ileal carcinoids with matching metastatic carcinoid to lymph nodes (n=5) and liver (n=1). The 30 formalin-fixed paraffin-embedded tissues used for ISH consisted of primary ileal carcinoids with matching liver metastases.
Expression of miRNAs in Primary and Metastatic Carcinoid Tumors
Analysis of the 95 miRNAs with real-time RT–PCR array showed that 10 miRNAs did not amplify consistency and were excluded form the study. Samples were normalized against the Ct value of miR-197, and the relative usage of miRNA expression was expressed in metastatic compared to the primary carcinoid tumors using the ΔΔCt method. Using Gene Spring Gx 10.0 software, we identified miRNAs with a metastatic/primary ratio less than 1 (upregulated in primary tumors) in all eight samples. The ranking of samples that were highest in the primary compared to the metastatic tumor (downregulated) is shown in Table 2. MiRNA-133a was ranked as one, whereas miRNA-145, -146, -222, and -106 were ranked lower (Figures 1a–d).
(a–d) Distribution of PCR array from eight ileal carcinoid tumors as log of fold change expressed as the ratio of metastatic to primary carcinoid tumors; 1 μg log of total RNA was used from each carcinoid tumor using a QuantiMir kit. Results were expressed relative to microRNA-197. Relative change of miRNA expression was calculated by the comparative CT method. (a–d) show the distribution as fold change for most of the miRNAs analyzed. Sixty-two microRNAs are labeled in the four figures.
The ranking of samples that were highest in the metastatic compared to the primary tumor (upregulated) are shown in Table 3 (Figures 1a–d). These included miRNA-183, -488, and -19a+b. The other miRNAs were more variably downregulated and upregulated between samples. A notable exception was miRNA122, which seemed to be highly upregulated in the metastatic tumors. However, as miRNA122 was highly expressed in liver tissue, this was interpreted to represent contaminated liver tissue in the metastatic cases.
Real-Time RT–PCR
Taq-Man RT–PCR analysis of miRNA-133a was performed with the eight original paired samples and after gel electrophoresis; the results were analyzed by densitometry (Figures 2a–d). There was a significant downregulation of miRNA-133a between the primary and metastatic carcinoids when samples were normalized in RNU48 from the same case. Comparison of the entire group (n=8) showed highly significant downregulation when quantitated by relative densitometry units (P<0.009). Two individual cases (cases 2 and 4) showed significant downregulation when the individual cases were evaluated. Analysis of six additional samples showed a similar trend in downregulation of miRNA-133a between the primary and metastatic tumors (P<0.014).
TaqMan qRT–PCR analysis for miRNA-133a followed by gel electrophoresis and densitometric analysis of eight matching primary and metastatic ileal carcinoid tumors (a–d). The upper panel shows the relative densitometric analysis of miR-133a relative to RNU48 for each sample, whereas the lower panels show the original gel samples. C1 through C8 are the RNU48 samples. P represents the primary tumors and M represents the metastatic samples. Results are mean±s.e. of the mean. *P<0.05 **P<0.001.
Analysis of Normal Enterochromaffin Cells
After laser capture microdissection and real-time RT–PCR, the enriched enterochromaffin cells were found to express relatively high levels of miRNA-133a when the results were normalized with RNU48 (Figure 3).
TaqMan qRT–PCR analysis for miRNA-133a followed by gel electrophoresis and densitometric analysis of four samples of enterochromaffin cells collected from 5 × 104 enterochromaffin cells. The upper panel shows the relative densitometric analysis of miR-133a relative to RNU48 for each of four experiments. C1 through C8 are the RNU48 samples and L represent the laser captured miRNA-133a samples. Results are expressed as mean±s.e. of the mean.
Northern Hybridization
Northern hybridization using matching pairs of primary and metastatic carcinoid tumors followed by densitometry analysis showed higher levels of miRNA-133a in primary tumors compared to metastatic tumors in two of the four samples analyzed when comparing miRNA-133a to RNU48 (Figure 4).
Northern hybridization analysis of four sets of matching primary and metastatic ileal carcinoid tumors. Total RNA of 20 μg was used for the northern hybridization with a Signos8is kit according to the manufacturer's protocol. The results were developed by enhanced chemiluminescence. Results were expressed relative to small nucleolar RNA, RNU48. RNA standards (20 nucleotides and 60 nucleotides) were run with the gel.
In Situ Hybridization
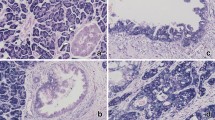
In situ hybridization using normal ileum showed expression of miRNA-133a in some of the enterochromaffin cells in the ileal mucosa (Figure 5a). ISH also showed expression of miRNA-133a in inflammatory cells including plasma cells and lymphocytes in the lamina propria of the ileum. The smooth muscle in the muscularis propria stained weakly, whereas the liver adjacent to hepatic metastases was negative for miRNA-133a. Analysis of 12 tumors with primary and metastasis using frozen tissue sections showed expression of miRNA-133a in both the primary and metastatic tumors (Figure 5b). Five cases showed higher levels of miRNA-133a in the primary tumors, three cases had higher levels in the metastases, whereas in four cases the levels were similar. The intensity of labeling between the primary and metastatic tumors was variable (Figures 5b and c). ISH analysis of miRNA-133a in 30 formalin-fixed paraffin-embedded tissue sections showed a positive hybridization signal in all cases (Figure 5C). Similar levels of staining intensity (1+ to 2+) were present in most cases from the formalin-fixed paraffin-embedded sections. In general, the hybridization signal was weaker in the formalin-fixed paraffin-embedded tissues compared to the frozen tissue sections. The control sections with a scramble control probe (Figure 5d) and after RNAse treatment (not shown) resulted in a negative miRNA signal.
In situ hybridization analysis using digoxigenin-labeled probes. (a) Normal ileal mucosa showing enterochromaffin cells with a positive hybridization signal for miRNA-133a (arrows). Formalin-fixed paraffin-embedded tissue sections; 200 nM of the 133a probe labeled at the 5′ and 3′ ends were used. (b) Frozen tissue sections of a primary ileal carcinoid tumor showing positive detection of miRNa-133a in the cytoplasm of tumor cells. The adjacent connective tissue is negative. (c) Paraffin section of a metastatic carcinoid tumor to the liver showing a strong positive hybridization signal for miRNA-133a in the cytoplasm of the tumor cells. The adjacent liver (L) is negative. (d) Paraffin section of hybridization with a scramble probe substituted for miRNA-133a used as a control. All other conditions were identical as with the miRNA-133a probe. There is no detection of miRNA-133a in the tumor or in the adjacent liver (L).
Discussion
Analysis of a unique 95 miRNA expression profile in primary and metastatic midgut carcinoid tumors showed consistent downregulation of miRNAs-133a, -145, -146, -222, and -10b in metastatic compared to primary carcinoids and upregulation of miRNA-183, -488, and -19a+b in metastatic carcinoid tumors. Validation of miRNA-133a in these neuroendocrine tumors was done by real-time RT–PCR, northern, and in situ hybridization.
MiRNA-133a has been studied in skeletal and cardiac muscle in which it has been found to be downregulated in hypertrophic muscle tissues.24, 25, 26, 27 MiRNA-133a has also been reported to be downregulated in squamous cell carcinoma of the tongue28 and miRNA-133a expression has been reduced in hepatocellular and lung carcinomas compared to normal tissues.29, 30 MiRNA-133a along with miRNA-1 are on chromosome 18 in the same bicistronic unit and are involved in muscle differentiation and proliferation.31
A recent study found that miRNA-133a was downregulated in neutrophils in myeloproliferative disorders.32 These findings would agree with our observation of miRNA-133a expression in inflammatory cells, including plasma cells and lymphocytes, in the lamina propria of the ileum by in situ hybridization. The mechanism of action of miR-133 on gene expression is currently being investigated. Xu et al33 reported that miRNA-133 and miR-1 had opposing effects on apoptosis, as miRNA-133 repressed caspase-9 expression at both the protein and mRNA level, whereas miRNA-1 reduced levels of HSP60 and H5P70 proteins without changing their transcription levels.
Several other miRNAs were consistently downregulated in metastatic carcinoid tumors compared to the primary tumors in this study. MiRNA-145 has been detected in various neoplasms.34, 35 It was reported to inhibit cell growth in some lung adenocarcinomas34 and is associated with decreased expression in gastric cancers.35 MiRNA-146 has been reported to suppress breast cancer metastasis.36 MiRNA-222 has been associated with various tumors including gliomas, prostate, and thyroid carcinomas.37, 38, 39 MiRNA-222 has been shown to target the cell cycle protein p27kip1 in various neoplasms.37, 38 MiRNA-10b is expressed in invasive gliomas and has also been associated with breast cancer metastasis.40, 41
Of the three miRNAs, which were most consistently upregulated in metastatic ileal carcinoids compared to the primary tumors, miRNA-183 has recently been shown to be overexpressed in metastatic colon cancer.42 An analysis of various miRNAs that are downregulated and/or upregulated in metastatic ileal carcinoid tumors may possibly help to predict the metastatic potential of these tumors.
These studies also show that the normal ileal enterochromaffin cells expresses miRNA-133a in both the primary and metastatic tumor. Densitometric analysis showed that the levels of expression by real-time RT–PCR were similar to that in the primary carcinoid tumors. However, as the laser captured enterochromaffin cells were mixed with ileal epithelial cells and constituted about 25% of the laser captured cells, we can assume that there may be at least 2- to 4-fold more miRNA-133a in the enterochromaffin cells compared to the primary tumor. If this assumption is correct, this would suggest that there is decreased expression of miRNA-133a in the development of primary carcinoids and a further decrease in miRNA-133a during progression from primary to metastatic carcinoid tumors. This suggests that decreasing levels of miRNA-133a has an important role in midgut carcinoid tumor development and progression.
This study also shows that in situ hybridization can be used to validate newly described miRNA in specific tumors and to show the localization of these miRNA in normal tissues as with miRNA-133a in the endocrine cells of the crypt epithelial cells in the ileum. Although in situ hybridization was useful in both frozen tissue sections and formalin-fixed paraffin-embedded tissues, higher probe concentrations and a longer development time to visualize the signal was needed with the formalin-fixed paraffin-embedded samples. We used probes labeled with digoxigenin at both the 3′ and 5′ ends to increase the detection signal, especially with formalin-fixed paraffin-embedded tissues. Although other studies have used locked nucleic acid probes labeled with digoxigenin to localize miRNA in formalin-fixed paraffin-embedded tissues,21, 22, 23 the use of probes labeled at both the 3′ and 5′ ends should further increase the sensitivity of the in situ hybridization procedure.
In summary, we have used a combination of RT–PCR arrays with the QuantiMir System to detect miRNAs in primary and metastatic ileal carcinoid tumors. MiRNA-133a was downregulated during progression from primary to metastatic carcinoid tumors. We used a combination of TaqMan real-time RT–PCR, northern, and in situ hybridization to demonstrate expression of miRNA-133a in carcinoid tumors and in enterochromaffin cells from the ileum. These findings suggest that miRNA-133a may have an important role in carcinoid tumor development and progression and that this miRNA may have diagnostic and/or prognostic utility in midgut carcinoid tumors.
References
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–866, review.
Esquela-Kerscher A, Slack FJ . Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–269.
Barbarotto E, Schmittgen TD, Calin GA . MicroRNAs and cancer: profile, profile, profile. Int J Cancer 2008;122:969–977.
Kent OA, Mendell JT . A small piece in the cancer puzzle; microRNAs as tumor suppressors and oncogenes. Oncogene 2006;25:6188–6196.
Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006;20:515–524.
Garson R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006;12:580–587.
Zhang W, Dahlberg JE, Tam W . MicroRNAs in tumorigenesis: a primer. Am J Pathol 2007;177:728–738.
Modin IM, Oberg K, Chung DC, et al. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol 2008;9:61–72.
Oberg K . Genetics and molecular pathology of neuroendocrine gastrointestinal and pancreatic tumors (gastroenteropancreatic neuroendocrine tumors. Curr Opin Endocrinol Diabetes Obes 2009;16:72–78.
Nilsson O, Wangberg B, McRae A, et al. Growth factors and carcinoid tumours. Acta Oncol 1993;32:115–124.
Oberg K . Expression of growth factors and their receptors in neuroendocrine gut and pancreatic tumors, and prognostic factors for survival. Ann N Y Acad Sci 2005;733:46–55.
Papouchado B, Erickson LA, Rohlinger AL, et al. Epidermal growth factor receptor and activated epidermal growth factor receptor expression in gastrointestinal carcinoids and pancreatic endocrine carcinomas. Mod Pathol 2005;18:1329–1335.
Stilling GA, Zhang H, Ruebel KH, et al. Characterization of the functional and growth properties of cell lines established from ileal and rectal carcinoid tumors. Endocr Pathol 2007;18:223–232.
Zhang HY, Rumilla KM, Jin L, et al. Association of DNA methylation and epigenetic inactivation of RASSF1A and beta catenin with metastasis in small bowel carcinoid tumors. Endocrine 2006;30:299–306.
Modlin IM, Kidd M, Pfragner R, et al. The functional characterization of normal and neoplastic human enterochromaffin cells. J Clin Endocrinol Metab 2006;91:2340–2348.
Modlin IM, Kidd M, Drozdov I, et al. Pharmacotherapy of neuroendocrine cancers. Expert Opin Pharmacother 2008;9:2617–2626.
Hobday TJ, Mahoney M, Erlichman C, et al. Preliminary results of a phase II trial of gefitinib in progressive metastatic neuroendocrine tumors (NET); A phase II consortium (P2C) study (Abstract 4083). J Clin Oncol 2005;23(part 1):328s.
Nakamura N, Ruebel K, Jin L, et al. Laser capture microdissection for analysis of single cells. Methods Mol Med 2007;132:11–18.
Leja J, Essaghir A, Essand M, et al. Novel markers for enterochromaffin cells and gastrointestinal neuroendocrine carcinomas. Mod Pathol 2009;22:261–272.
Riss D, Jin L, Qian X, et al. Differential expression of galectin-3 in pituitary tumors. Cancer Res 2003;63:2251–2255.
Nelson PT, Baldwin DA, Kloosteman WP, et al. Rake and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 2006;12:187–191.
Thomsen R, Nielsen PS, Jensen TH . Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA 2005;11:1745–1748.
Nuovo GJ . In situ detection of precursor and mature microRNAs in paraffin embedded, formalin-fixed tissues and cell preparations. Methods 2008;44:39–46, review.
Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006;38:228–233.
Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618.
Luo X, Lin H, Pan Z, et al. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem 2008;283:20045–20052.
McCarthy JJ, Esser KA . MicroRNA-1 and microRNA–133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 2007;102:306–313.
Wong TS, Liu XB, Chung-Wai HA, et al. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J Cancer 2008;123:251–257.
Datta J, Kutay H, Nasser MW, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res 2008;68:5049–5058.
Nasser MW, Datta J, Nuovo G, et al. Down-regulation of micro-RNa-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem 2008;283:33394–33405.
Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004;5:R13–R13.11.
Slezak S, Jin P, Caruccio L, et al. Gene and microRNa analysis of neutrophils from patients with polycythemia vera and essential thrombocytosis: down-regulation of micro RNA-1 and -133a. J Transl Med 2009;7:39, PubMed, in process.
Xu C, Lu Y, Pan Z, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 2007;120:3045–3052.
Cho WC, Chow AS, Au JS . Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 2009;45:2197–2206.
Takagi T, Ilio A, Nakagawa Y, et al. Decreased expression of MicroRNA-143 and -145 in human gastric cancers. Oncology 2009;77:12–21.
Hurst DR, Edmonds MD, Scott GK, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 2009;69:1279–1283.
Zhang C, Kang C, You Y, et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol 2009;34:1653–1660.
Schwertheim S, Sheu SY, Worm K, et al. Analysis of deregulated miRNAs is helpful to distinguish poorly differentiated thyroid carcinoma from papillary thyroid carcinoma. Horm Metab Res 2009;41:475–481.
Sun T, Wang Q, Balk S, et al. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cacner Res 2009;69:3356–3363.
Sasayama T, Nishihara M, Kondoh T, et al. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 2009;125:1407–1413.
Ma L, Teruya-Feldstein J, Weinberg RA . Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682–688.
Motoyama K, Inoue H, Takatsuno Y, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 2009;34:1069–1075.
Acknowledgements
The project described was supported by Grant Number 1 UL1 RR024150 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH. Information on NCRR is available at http://www.ncrr.hig.gov/. Information on Reengineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nig.gov. Supported in part by a Novel Methodology Grant from CTSA and by a grant from the Sackler Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Ruebel, K., Leontovich, A., Stilling, G. et al. MicroRNA expression in ileal carcinoid tumors: downregulation of microRNA-133a with tumor progression. Mod Pathol 23, 367–375 (2010). https://doi.org/10.1038/modpathol.2009.161
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.161
Keywords
This article is cited by
-
Molecular Pathology of Well-Differentiated Gastro-entero-pancreatic Neuroendocrine Tumors
Endocrine Pathology (2021)
-
Exosomal miRNA signatures of pancreatic lesions
BMC Gastroenterology (2020)
-
Treatment with somatostatin analogs induces differentially expressed let-7c-5p and mir-3137 in small intestine neuroendocrine tumors
BMC Cancer (2019)
-
Methylation-mediated silencing of miR-133a-3p promotes breast cancer cell migration and stemness via miR-133a-3p/MAML1/DNMT3A positive feedback loop
Journal of Experimental & Clinical Cancer Research (2019)
-
Role of biomarker tests for diagnosis of neuroendocrine tumours
Nature Reviews Endocrinology (2018)