Abstract
Solid pseudopapillary neoplasms of the pancreas are low-grade malignancies, but their biological behavior cannot be stratified solely on the basis of histopathologic criteria. Aside from mutations in β-catenin and lack of genetic changes common to pancreatic ductal adenocarcinomas, little is known about the chromosomal alterations in solid pseudopapillary neoplasms. We applied array comparative genomic hybridization to a series of 12 patients. The average age was 31 years (range 12–52 years) with 10 female and 2 male patients. The average tumor size was 7.3 cm (range 2–24 cm) with five lesions greater than 5 cm. All cases had ‘bland’ cytology without significant pleomorphism or high nuclear grade, but seven cases demonstrated at least one of these potentially aggressive histopathologic features: size >5 cm, tumor necrosis, lymphovascular invasion, perineural invasion and peripancreatic invasion. Clinically, one lesion demonstrated aggressive behavior. By array comparative genomic hybridization, chromosomal losses and/or gains were identified in eight cases; five cases had multiple (five or more) alterations. The most common alterations were gains at 13q, 17q, 1q and 8q. Six of the seven cases with at least one aggressive feature had genetic alterations, while only two cases without adverse features had genetic alterations (P=0.024). The single clinically aggressive tumor exhibited seven chromosomal gains and four aggressive histopathologic features. Our study demonstrates that genetic alterations detected by array comparative genomic hybridization are common in solid pseudopapillary neoplasms of the pancreas. Additional study and longer follow-up are needed to determine if these genetic abnormalities could help predict clinical behavior in these neoplasms.
Similar content being viewed by others
Main
Solid pseudopapillary neoplasm of the pancreas is a relatively rare neoplasm that tends to affect young women. These tumors are considered to be low-grade malignancies and complete resection is usually curative. However, aggressive malignant solid pseudopapillary neoplasms have been reported.1, 2, 3, 4, 5, 6 Attempts to separate aggressive from non-aggressive solid pseudopapillary neoplasms based on histologic criteria (such as vascular invasion, necrosis, increased mitotic rate, high nuclear grade) and size has yielded contradictory results with reports of histologically bland solid pseudopapillary neoplasms that have metastasized and histologically malignant solid pseudopapillary neoplasms with indolent outcomes.4, 7, 8 Using morphologic features to predict the biological behavior of solid pseudopapillary neoplasms has limitations and is a major obstacle in stratifying patients into low- and high-risk groups as has been observed for other lesions, such as pheochromocytomas and neuroendocrine tumors.
Recent studies have applied cytogenetic and molecular analysis to solid pseudopapillary neoplasms. Both aneuploid and diploid tumors have been detected by flow cytometry.3, 6, 8, 9, 10, 11 Various chromosomal abnormalities have also been reported, primarily using cytogenetic techniques.12, 13, 14, 15, 16 Two studies using comparative genomic hybridization (CGH) found no chromosomal gains or losses,11, 17 while in another study of one case using the more sensitive array CGH technique, two alterations were detected: loss of heterozygosity for HRAS in chromosome band 11p15.5 and a less significant loss of the short arm of chromosome 16.13 Unlike ductal adenocarcinomas of the pancreas, solid pseudopapillary neoplasms do not typically have KRAS, p16, DPC4 or p53 gene alterations but do harbor mutations in the APC/β-catenin pathway with overexpression of β-catenin and cyclin D1.18 Despite early attempts to elucidate the molecular characteristics of solid pseudopapillary neoplasms, the role of molecular analysis in risk stratification for individuals and the morphologic correlations of these genetic changes are unknown. We therefore undertook genome-wide evaluation of a series of 12 solid pseudopapillary neoplasms by array CGH and correlated genetic alterations with aggressive tumor characteristics.
Materials and methods
Case Selection
Twelve solid pseudopapillary neoplasms with available archived tissue (formalin-fixed, paraffin-embedded blocks) were retrieved from University of Pittsburgh pathology files between November 2003 and December 2006: 10 specimens were from adult patients and 2 were from children. The clinicopathologic data from each case were tabulated. Array CGH was performed with fluorescence in situ hybridization validation. Study procedures were approved by the Institutional Review Board.
Array Comparative Genomic Hybridization
Gene gains and losses were detected by the commercially available VYSIS® GenoSensor™ Array 300 genomic DNA microarray kit (Abbott Molecular, Des Plaines, IL, USA) that contains triplicates of 287 target clone DNAs (P1 or BAC clones) representing oncogenes and tumor suppressor genes. The target tissue was microdissected from ten 4-μm-thick unstained histologic sections under direct visualization using a stereoscopic microscope. Genomic DNA was extracted by proteinase K digestion and DNEasy DNA extraction column (Qiagen, Valencia, CA, USA). The DNA concentration was quantified using a fluorospectrometer (Nanodrop, Wilmington, DE, USA). DNA quality was assured on 2% agarose gel.
DNA labeling and hybridization were performed according to the manufacturer's instructions. In brief, tumor DNA and reference DNA were labeled by random priming reaction (Random Priming Reaction Kit, Vysis, Downers Grove, IL, USA) with Cy3-dCTp and Cy5-dCTP (Perkin Elmer Life Sciences Inc., Boston, MA, USA), respectively. Labeled tumor DNA and reference DNA were mixed with Microarray Hybridization Buffer (Vysis) containing Cot-1 DNA, followed by denaturation at 80°C for 10 min, followed by 1 h of incubation at 37°C. Thirty microliters of hybridization mixture was transferred onto the GenoSensor Array 300 microarray template (Abbott Molecular). Hybridization was carried out for 7 days at 37°C. Post-hybridization washes were performed using washing solution (2 × SSC/50% formamide) at 40°C (3 × 10 min), followed by 1 × SSC (4 × 5 min) and a 1–2 s rinse in distilled water. Hybridized DNA was counterstained with DAPI IV solution (Vysis).
The hybridized microarray slides were analyzed using the GenoSensor Reader System (Abbott Molecular). Cy3/Cy5 ratios were automatically determined for each target. The normalized ratio of the test DNA copy number relative to the normal reference DNA copy number was calculated. According to the manufacturer's instructions and our own validation using normal DNA vs normal reference DNA, the cutoff fluorescence ratio between normal and aberrant DNA copy numbers was at mean 1.00±2 s.d. Fluorescence ratios ≥1.2 were considered as DNA sequence copy number gains and fluorescence ratios ≤0.80 were considered as DNA sequence copy number losses. Statistical significance of chromosomal copy number change was determined at P<0.0001.
Fluorescence In Situ Hybridization
Gains at 13q14 were validated by fluorescence in situ hybridization on sequential sections of the same tumor blocks. Dual-color fluorescence in situ hybridization was performed on deparaffinized slides using the LSI 13 (13q14) RB1 SpectrumOrange probe and the LSI 13q34 SpectrumGreen probe (Vysis). Briefly, slides were deparaffinized, dehydrated, pretreated with the Vysis Paraffin Pretreatment Kit (Vysis) and digested for 18 min in protease solution. The slides were incubated with probe, which was denatured prior to hybridization, overnight at 37°C in a humidified chamber. Post-hybridization, the slides were washed, air-dried in the dark and counterstained with DAPI. Analysis was performed using a Nikon Optiphot-2 and Quips Genetic Workstation equipped with Chroma Technology 83 000 filter set with single band exitors for Texas Red/rhodamine, FITC and DAPI (UV 360 nm). Only individual and well-delineated cells were scored. Overlapping cells were excluded from the analysis. Approximately 60 cells were analyzed in the targeted region.
Statistical Analysis
Nonparametric statistical analysis was performed to correlate the frequency of chromosomal aberrations with the presence of adverse histologic features (Statview, SAS Institute).
Results
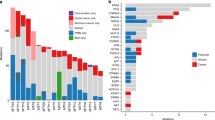
The clinicopathologic data are summarized in Table 1. The average age at presentation was 31 years (range 12–52 years) with a 10:2 female/male ratio. Five patients presented with abdominal pain. Six presented with unrelated medical conditions (eg upper gastrointestinal bleed) and one was asymptomatic. The solid pseudopapillary neoplasms were located in the head (two cases), body (four cases) and tail (five cases) of the pancreas, and in one case (case 10), replaced the entire pancreas, invaded into the stomach and spread into the peritoneal cavity. The average tumor size was 7.3 cm (range 2–24 cm) with five lesions larger than 5 cm. None of the cases had positive lymph nodes. Microscopically, classic features of solid pseudopapillary neoplasms were seen in all cases, including discohesive, monotonous cells with papillae, foamy macrophages and cholesterol clefts, dense hyaline bands and hyaline globules. One lesion (case 10) was clinically aggressive. In this case, the patient presented with disseminated peritoneal disease and hepatic metastases. The majority of the peritoneal tumor was removed following two debulking procedures, but the liver metastases were not resected. The patient received chemotherapy and additional peritoneal tumor was removed 8 months later. The patient's most recent biopsy was taken 1 month prior to the end of this study, 15 months after the initial presentation, and showed viable tumor in the liver. None of the remaining cases have recurred to date, with an average follow-up of 33 months (range 12–50 months).
Based on a literature search, we selected the following histopathologic features as indicators of potentially aggressive tumor biology: size >5 cm,7 tumor necrosis (‘necrobiotic nests’),4, 8 invasion into peripancreatic soft tissue,7 lymphovascular/perineural invasion,7, 8 increased mitotic/proliferative rate4, 7, 8 and a high degree of cellular pleomorphism.8 Seven patients demonstrated at least one aggressive feature (Table 1). Five tumors were >5 cm; true tumor necrosis characterized by necrobiotic nests was present in one case (Figure 1a), lymphovascular invasion in two cases (Figure 1b), perineural invasion in three cases (Figure 1c) and peripancreatic invasion in one case. None of the cases had increased mitotic activity; 10 cases had 0 mitosis/50 hpf and two cases had 1 mitosis/50 hpf; immunohistochemistry for the proliferation marker ki-67 revealed a low proliferation rate of 2% or less in all tumors. None of the cases showed significant cellular pleomorphism or high nuclear grade.
Aggressive histologic features seen in our cases. (a) One tumor exhibited true tumor necrosis with ‘necrobiotic nests.’ (b) Lymphovascular invasion was seen in two cases. (c) Perineural invasion was seen in three cases (‘N’=nerves). Peripancreatic invasion was present in one case, but this was best observed grossly.
By array CGH, chromosomal abnormalities were identified in eight (67%) of the cases (Table 2). The most common gains involved 13q (five cases), 17q (five cases), 1q (three cases) and 8q (three cases); the chromosomal gains at 13q14 were confirmed by fluorescence in situ hybridization (Figure 2). Chromosomal losses were infrequent but losses in 11q were detected in two cases. Of the five cases with the most chromosomal alterations (five or more, cases 8–12), all (100%) had indicators of aggressive behavior: four had aggressive histologic features and one was a 9.9 cm tumor (Tables 1 and 2). Statistical analysis demonstrated a significant correlation between the presence of one or more aggressive feature and the frequency of chromosomal abnormalities (P=0.024, Mann–Whitney U-test). Of the seven cases with aggressive features, six (86%) cases had genetic alterations: five cases showed at least five alterations and one case had no chromosomal alterations (Table 2). Correspondingly, the single clinically aggressive lesion (case 10) exhibited seven chromosomal gains and displayed four histopathologic criteria predictive of aggressive tumor biology, including tumor size of 24 cm, peripancreatic invasion, perineural invasion and lymphovascular invasion.
Discussion
Solid pseudopapillary neoplasm of the pancreas is an uncommon neoplasm observed primarily in young women. These tumors are usually found incidentally and are treated by pancreatic resection. The morphologic features of solid pseudopapillary neoplasms are distinct. Although generally considered low-grade neoplasms, solid pseudopapillary neoplasms can exhibit aggressive tumor biology. The histologic characteristics of solid pseudopapillary neoplasms tend to have less prognostic significance than the clinicopathologic characteristics of the neoplasm, such as tumor size and extent of disease, as is observed for other tumors, such as pheochromocytomas, gastrointestinal stromal tumors and neuroendocrine tumors.
The morphologic features suggesting a benign or malignant phenotype are inadequate to predict tumor biology accurately.1, 2, 3, 4, 5, 6, 7, 8 The application of simple DNA-based ancillary techniques has yielded mixed results as applied to high- and low-risk solid pseudopapillary neoplasms. Flow cytometry studies have demonstrated a trend toward DNA aneuploidy in malignant solid pseudopapillary neoplasms, but diploidy has been observed in both malignant and ‘benign’ solid pseudopapillary neoplasms.3, 6, 8, 9, 10, 12 Several case reports using classic cytogenetics have found significant abnormalities in solid pseudopapillary neoplasms. One tumor that demonstrated aggressive tumor biology with local invasion into the bile duct as well as high mitotic activity had a karyotypic profile of double loss of X chromosomes and trisomy 3.15 In contrast, a more recent pediatric solid pseudopapillary neoplasm with low-risk morphology (bland morphology and no perineural, vascular or pancreatic invasion, but 8.5 cm in size with areas of necrosis) revealed two clones, one with complex karyotypic changes that involved four translocations and a duplication and another with partial monosomy for chromosome X.13 Additional karyotyping studies have been similarly contradictory. A 19 cm solid pseudopapillary neoplasm with bland morphologic features had a dramatically abnormal karyotype (45,XX,−13,der(17)t(13;17)(q14;p11)(12)) and a small (5 cm) lesion with areas of necrosis but otherwise benign features had a complex karyotype defined as 46,XX, der(1)add(1)(p?)(q?), del(14)(q22), der(20)t(1;20)(q21;q13).16
Studies using more sophisticated techniques have revealed additional genetic alterations in solid pseudopapillary neoplasms. Solid pseudopapillary neoplasms are now known to harbor an exon 3 mutation in the β-catenin gene,18, 19 which causes β-catenin overexpression. In addition to β-catenin mutations, an EWS/FLI-1 fusion transcript, t(11;22)(q24;q12), has been reported in a pediatric solid pseudopapillary neoplasm.14 Subsequent papers showed that although FLI-1 is overexpressed in solid pseudopapillary neoplasms,17 it is more likely due to a genetic alteration involving the long arm of chromosome 11 rather than an EWS-FLI-1 transcript.20 Interestingly, two of our patients showed loss of 11q22.3, and one showed a gain at 11q13.5–q14, suggesting a role for changes in chromosome 11q during the development of solid pseudopapillary neoplasms.
Published reports using CGH to analyze solid pseudopapillary neoplasms have demonstrated mixed results. One study of 30 solid pseudopapillary neoplasms and an additional case report did not show any chromosomal loss or gain.11, 17 Interestingly, all these cases had benign features. Using the more sensitive array CGH method, a recent report demonstrated loss of heterozygosity for HRAS in chromosome band 11p15.5 and a less significant loss of the short arm of chromosome 16.13 This case also had cytogenetic analysis and had potentially aggressive features including size greater than 5 cm and areas of necrosis.13
Our array CGH analysis identified numerous chromosomal abnormalities in a small series of 12 solid pseudopapillary neoplasms. Some tumors had no detectable alterations, while others had several. Statistical analysis demonstrated an association between ‘aggressive’ tumor characteristics including malignant biology and the number of chromosomal alterations. Although prior studies using less sensitive non-array CGH methods have identified no genetic abnormalities, we used stringent cutoff values for statistical significance in the analysis of the gene array data. Furthermore, many of the alterations encountered in this series have been reported previously using different methods. We conclude that chromosomal abnormalities detected by array CGH are common in solid pseudopapillary neoplasms of the pancreas. The significance of changes at 11q, 13q, 17q, 1q and 8q require further study. Longer follow-up and further correlation with recurrence rates and survival in our patients may lead to molecular prognostication in solid pseudopapillary neoplasms.
References
Alexandrescu DT, O'Boyle K, Feliz A, et al. Metastatic solid-pseudopapillary tumour of the pancreas: clinico-biological correlates and management. Clin Oncol (R Coll Radiol) 2005;17:358–363.
Hibi T, Ojima H, Sakamoto Y, et al. A solid pseudopapillary tumor arising from the greater omentum followed by multiple metastases with increasing malignant potential. J Gastroenterol 2006;41:276–281.
Lai HW, Su CH, Li AF, et al. Malignant solid and pseudopapillary tumor of the pancreas—clinicohistological, immunohistochemical, and flow cytometric evaluation. Hepatogastroenterology 2006;53:291–295.
Tang LH, Aydin H, Brennan MF, et al. Clinically aggressive solid pseudopapillary tumors of the pancreas: a report of two cases with components of undifferentiated carcinoma and a comparative clinicopathologic analysis of 34 conventional cases. Am J Surg Pathol 2005;29:512–519.
Choi SH, Kim SM, Oh JT, et al. Solid pseudopapillary tumor of the pancreas: a multicenter study of 23 pediatric cases. J Pediatr Surg 2006;41:1992–1995.
Kamei K, Funabiki T, Ochiai M, et al. Three cases of solid and cystic tumor of the pancreas. Analysis comparing the histopathological findings and DNA histograms. Int J Pancreatol 1991;10:269–278.
Kang CM, Kim KS, Choi JS, et al. Solid pseudopapillary tumor of the pancreas suggesting malignant potential. Pancreas 2006;32:276–280.
Nishihara K, Nagoshi M, Tsuneyoshi M, et al. Papillary cystic tumors of the pancreas. Assessment of their malignant potential. Cancer 1993;71:82–92.
Cho NH, Go JH, Jung SH, et al. Correlation between proliferating index and prognostic factors in papillary cystic tumors of the pancreas. J Korean Med Sci 1995;10:342–351.
Pettinato G, Manivel JC, Ravetto C, et al. Papillary cystic tumor of the pancreas. A clinicopathologic study of 20 cases with cytologic, immunohistochemical, ultrastructural, and flow cytometric observations, and a review of the literature. Am J Clin Pathol 1992;98:478–488.
Tornoczky T, Kalman E, Jakso P, et al. Solid and papillary epithelial neoplasm arising in heterotopic pancreatic tissue of the mesocolon. J Clin Pathol 2001;54:241–245.
Grant LD, Lauwers GY, Meloni AM, et al. Unbalanced chromosomal translocation, der(17)t(13;17)(q14;p11) in a solid and cystic papillary epithelial neoplasm of the pancreas. Am J Surg Pathol 1996;20:339–345.
Kempski HM, Austin N, Chatters SJ, et al. Previously unidentified complex cytogenetic changes found in a pediatric case of solid-pseudopapillary neoplasm of the pancreas. Cancer Genet Cytogenet 2006;164:54–60.
Maitra A, Weinberg AG, Schneider N, et al. Detection of t(11;22)(q24;q12) translocation and EWS-FLI-1 fusion transcript in a case of solid pseudopapillary tumor of the pancreas. Pediatr Dev Pathol 2000;3:603–605.
Matsubara K, Nigami H, Harigaya H, et al. Chromosome abnormality in solid and cystic tumor of the pancreas. Am J Gastroenterol 1997;92:1219–1221.
Stringer MD, Roberts P, Davison SM, et al. A novel cytogenetic abnormality in a solid and cystic papillary tumour of the pancreas. Med Pediatr Oncol 2003;41:155–158.
Tiemann K, Kosmahl M, Ohlendorf J, et al. Solid pseudopapillary neoplasms of the pancreas are associated with FLI-1 expression, but not with EWS/FLI-1 translocation. Mod Pathol 2006;19:1409–1413.
Abraham SC, Klimstra DS, Wilentz RE, et al. Solid-pseudopapillary tumors of the pancreas are genetically distinct from pancreatic ductal adenocarcinomas and almost always harbor beta-catenin mutations. Am J Pathol 2002;160:1361–1369.
Tanaka Y, Kato K, Notohara K, et al. Frequent beta-catenin mutation and cytoplasmic/nuclear accumulation in pancreatic solid-pseudopapillary neoplasm. Cancer Res 2001;61:8401–8404.
Tiemann K, Heitling U, Kosmahl M, et al. Solid pseudopapillary neoplasms of the pancreas show an interruption of the Wnt-signaling pathway and express gene products of 11q. Mod Pathol 2007;20:955–960.
Acknowledgements
We thank Kathleen Cieply, Tracy Mercuri, Carol Sherer and Kathleen Cumbie in the Fluorescence In Situ Hybridization Laboratory of the Department of Pathology, University of Pittsburgh Medical Center, for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rund, C., Moser, A., Lee, K. et al. Array comparative genomic hybridization analysis of solid pseudopapillary neoplasms of the pancreas. Mod Pathol 21, 559–564 (2008). https://doi.org/10.1038/modpathol.2008.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.6
Keywords
This article is cited by
-
Solid pseudopapillary tumor of the pancreas: The surgical procedures
Surgery Today (2011)