Abstract
The distinction between chondrosarcoma and chordoma of the skull base/head and neck is prognostically important; however, both have sufficient morphologic overlap to make delineation difficult. As a result of gene expression studies, additional candidate markers have been proposed to help in separating those entities. We sought to evaluate the performance of new markers: brachyury, SOX-9, and podoplanin alongside the more traditional markers glial fibrillary acid protein, carcinoembryonic antigen, CD24, and epithelial membrane antigen. Paraffin blocks from 103 skull base/head and neck chondroid tumors from 70 patients were retrieved (1969–2007). Diagnoses were made based on morphology and/or whole-section immunohistochemistry for cytokeratin and S100 protein yielding 79 chordomas (comprising 45 chondroid chordomas and 34 conventional chordomas), and 24 chondrosarcomas. A tissue microarray containing 0.6 mm cores of each tumor in triplicate was constructed using a manual array (MTA-1; Beecher Instruments). For visualization of staining, the ImmPRESS detection system (Vector Laboratories) with 2-diaminobenzidine substrate was used. Sensitivities and specificities were calculated for each marker. Core loss from the microarray ranged from 25 to 29% yielding 66–78 viable cases per stain. The classic marker, cytokeratin, still has the best performance characteristics. When combined with brachyury, accuracy improves slightly (sensitivity and specificity for detection of chordoma 98 and 100%, respectively). Positivity for both epithelial membrane antigen and AE1/AE3 had a sensitivity of 90% and a specificity of 100% for detecting chordoma in this study. SOX-9 is apparently common to both notochordal and cartilaginous differentiation, and is not useful in the chordoma-chondrosarcoma differential diagnosis. Glial fibrillary acid protein, carcinoembryonic antigen, CD24, and epithelial membrane antigen did not outperform other markers, and are less useful in the diagnosis of chordoma vs chondrosarcoma. Podoplanin still remains the only positive marker for chondrosarcoma, though its accuracy is less than previously reported.
Similar content being viewed by others
Main
Chordomas and chondrosarcomas represent two morphologically similar, but biologically distinct categories of mesenchymal neoplasms that can involve the skull base. Chordomas are rare, slow growing tumors derived from remnant notochord occurring anywhere along the central neural axis. In the adult, an estimated 33–37% are located at the skull base.1, 2, 3, 4, 5 Chondrosarcomas are rare cartilaginous malignancies comprising 6–15% of all skull base tumors.2, 3, 4, 6, 7 The vast majority of skull base chondrosarcomas are well differentiated; high-grade and dedifferentiated chondrosarcomas are virtually nonexistent. Chondroid chordoma is a histologic variant of chordoma that has a predilection for the skull base and is defined by the presence of varying amounts of cartilaginous elements admixed with cords and lobules of vacuolated or ‘physaliferous’ cells typical of conventional chordoma. Originally described by Heffelfinger et al5 in 1973, chondroid chordomas have sparked debate over the past three decades regarding their histogenesis, accurate diagnosis, and prognosis.8, 9, 10, 11, 12, 13
Current evidence suggests that chondroid chordomas are indeed variants of chordomas with outcomes similar to those of conventional chordomas.14, 15, 16, 17, 18 However, the histologic similarities between chondroid chordoma and chondrosarcoma often still pose diagnostic challenges, especially on small biopsies. Both are treated similarly, but, in spite of the introduction of newer radiotherapy-based modalities, chondroid chordoma retains a worse prognosis compared to chondrosarcoma in this region.14, 19, 20 A variety of immunohistochemical markers have been used to better delineate these entities. Historically, the selection of these discriminatory markers has been modeled after the expression profile of notochord.21, 22, 23, 24 Of these, the most effective and well-established markers selectively expressed in chordomas, but not chondrosarcomas, are cytokeratin, followed by epithelial membrane antigen (EMA).13, 25, 26, 27 Although these markers have performed well in the literature, a panel-based approach incorporating multiple markers may provide a more robust confirmation of diagnosis—especially if one stain is equivocal or technically compromised.15, 28 Other markers such as carcinoembryonic antigen (CEA) and glial fibrillary acidic protein (GFAP) have shown far less utility in distinguishing chordoma from chondrosarcoma.29, 30, 31
Recently, gene expression profiling has expanded the list of possible discriminatory markers. Among those selectively expressed in chordoma are CD24 and brachyury.16, 32, 33 CD24 is a glycoprotein that functions in cell adhesion and has been shown to be expressed by the notochord-derived nucleus pulposus.33 Brachyury is the transcription factor protein product of a T-box gene whose function is to regulate formation of the mesoderm and notochord in humans.34 Immunohistochemical studies demonstrated that brachyury expression is highly sensitive and specific for chordomas.16 In fact, because of its initial performance, brachyury has recently been proposed as the ultimate solution to this differential diagnosis.15
The bulk of investigative efforts in this differential diagnosis have focused on markers that are selectively expressed in chordoma. But recently, Huse et al28 have discovered the first reliable immunohistochemical marker that is selectively expressed in chondrosarcomas, namely, D2-40, a monoclonal antibody that recognizes the O-linked sialoglycoprotein, podoplanin. However, the performance of this marker has not been evaluated on a large series of tumors. Another purported chondrogenesis-selective marker is SOX-9. SOX-9 is a homeobox transcription factor that is heavily involved in chondrogenesis in mammals.35, 36 SOX-9 expression has been identified in chondrosarcomas37 and has been used in distinguishing mesenchymal chondrosarcoma from other round blue cell tumors.38 However, SOX-9 is also vital to other organ development and is expressed in other tumor types as well, suggesting that its utility would be limited to specific diagnostic considerations.39, 40 To date, this marker has not been evaluated with respect to the chordoma-chondrosarcoma differential diagnosis.
Although many of these newer markers are promising, they have not been compared systematically to each other or to the more traditional markers used in the differential diagnosis. To evaluate the performance characteristics of several traditional and newer markers, we used tissue microarray methodology to assess a large series of chordomas and chondrosarcomas of the skull base.
Materials and methods
Case Selection
This study was approved by our institutional review board (no. 0701113). Paraffin blocks and slides from 103 skull base/head and neck tumors from 70 patients were successfully retrieved from the University of Pittsburgh Medical Center Department of Pathology Archives (1969–2007). The tumors consisted of 79 chordomas (45 chondroid chordomas, 34 conventional chordomas), and 24 chondrosarcomas (23 grades 1–2, 1 grade 3). Three chondrosarcomas also showed at least focal dedifferentiation. One chondrosarcoma was seen in the setting of Mafucci syndrome. In 32% (25 of 79) of the chordomas, solid cellular areas consisted greater than 5% of the tumor. The original whole-section slides and/or recuts and immunohistochemical stains were verified by two observers (GJO and RRS) to establish gold standard diagnoses. For 63 of the 70 patients clinical follow-up data were available. Median follow-up on surviving patients was 85 months (range: <1–230 months). Patient outcome characteristics in our series are summarized in Table 1.
Tissue Microarray Construction and Immunohistochemistry
Paraffin ‘donor’ blocks were selected for each case. Using a manual tissue arrayer (MTA-1; Beecher Instruments, Sun Prairie, WI, USA) 0.6 mm cores were transferred from each donor block to a blank recipient paraffin block and arrayed in triplicate by one of the authors (RRS). Use of decalcified blocks could not be avoided in 65% (66 of 102) of our cases. For chondroid chordomas, whenever possible, at least one core was obtained from chondroid predominant areas, and one core from more conventional appearing areas. The cores from the donor block were annealed to the recipient block by heating to 37°C for 5 min and gently pressing on a flat surface to make all cores level. Paraffin sections from the constructed tissue microarray block were cut (without tape transfer) and incubated with commercially available antibodies for S100 protein, cytokeratin cocktail AE1/AE3, brachyury, EMA, CD24, podoplanin, GFAP, polyclonal CEA, and SOX-9. Antibodies, dilutions, company, and pretreatment are summarized in Table 2. Staining was visualized using the ImmPRESS™ (Vector Laboratories, Burlingame, CA, USA) detection system with 2-diaminobenzidine as the substrate chromogen (Table 3).
Data Analysis
Each antibody was scored manually by two observers (GJO and RRS). A core was considered positive if at least 10% of the core showed staining in the expected cell compartment distribution. A case was considered positive if at least one core was positive. Discrepancies between observers were resolved by simultaneous review. Sensitivities and specificities were calculated using Microsoft Excel 2003, version 11.6355.6360 (Microsoft Corp). Survival analysis was performed using the Kaplan–Meier method with SPSS software, version 14.0 (SPSS, Chicago, IL, USA). Group-wise comparisons were made using the log-rank test.
Results
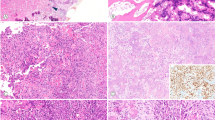
Whole sections representing chondrosarcoma, conventional chordoma, and chondroid chordoma are shown in Figure 1. Core loss from the microarray ranged from 25 to 29% yielding 66–78 viable cases per stain. The immunoprofiles based on the remaining cores are summarized in Tables 3 and 4. Examples of observed staining patterns for the diagnoses and immunohistochemical stains used are illustrated in Figure 2.
Observed immunohistochemistry for microarray cores of chordoma and chondrosarcoma for the following markers: S100 protein, cytokeratin AE1/AE3, brachyury, podoplanin, EMA, CD24, SOX-9, GFAP, and polyclonal CEA (original magnification × 100). Abbreviations: AE1/3, cytokeratin AE1/AE3; Brachy, brachyury; Podo, podoplanin.
Classic Markers
All cores stained for S100 protein, comprising 23 conventional chordomas, 33 chondroid chordomas, and 18 chondrosarcomas. Briefly, 25 of 25 (100%) of conventional chordomas, 32 of 34 (94%) of chondroid chordomas, and 0 of 18 (0%) of chondrosarcomas were stained with AE1/AE3. With EMA, 19 of 20 (95%) of conventional chordoma, 21 of 23 (91%) of chondroid chordoma, and 4 of 16 (25%) of chondrosarcoma were positive. Polyclonal CEA showed positive staining in only 8 of 24 (33%) of conventional chordoma, 9 of 31 (29%) of chondroid chordoma, and 0 of 18 (0%) of chondrosarcoma. Using GFAP, only 8 of 22 (36%) of conventional chordoma, 18 of 33 (55%) of chondroid chordoma, and 1 of 16 (6%) of chondrosarcoma were positive.
Newer Markers
Of the newer markers that are selective for chordoma, brachyury performed best, staining 22 of 25 (88%) of conventional chordomas, 29 of 32 (91%) of chondroid chordomas, and 0 of 16 (0%) of chondrosarcomas. CD24 staining showed that only 11 of 23 (48%) of conventional chordomas, 16 of 35 (46%) of chondroid chordomas, and 0 of 14 (0%) of chondrosarcomas were positive. Of the chondrosarcoma-selective markers, podoplanin was positive in 14 of 18 (78%) of chondrosarcomas, 1 of 21 (5%) of conventional chordomas, and 7 of 33 (21%) of chondroid chordomas. However, SOX-9 staining was virtually ubiquitous; positive in 16 of 17 (94%) of chondrosarcomas, but also 24 of 25 (96%) of conventional chordomas and 34 of 34 (100%) of chondroid chordomas.
Unusual Cases Including the Brachyury-Negative Chordomas
On the tissue microarray, one chordoma was actually negative for cytokeratin AE1/AE3, but positive for brachyury and S100 protein (Figure 3). The whole-mount slides of this particular chordoma showed rare solid areas and approximately 10% chondroid differentiation, with the initial report describing only focal pancytokeratin reactivity. Eight chordomas were positive for podoplanin, including seven chondroid and one conventional chordomas. These eight chordomas were not histologically distinct from any of the other chordomas, although all cases showed some myxoid and/or solid areas. These eight podoplanin-positive chordomas were also positive for cytokeratin AE1/AE3 (Figure 4). The brachyury-negative, cytokeratin-positive chordomas consisted of three chondroid chordomas, and four predominantly solid chordomas comprised of more atypical cells with mitoses and necrosis, including one ‘rhabdoid’ chordoma (Figure 5). Interestingly, within chordomas, brachyury negativity also showed a trend toward poorer overall survival (P-value 0.099; Figure 6). Other correlations between immunomarkers and outcome within chordomas and chondrosarcomas were not significant (data not shown).
The sole chordoma that was not positive for AE1/AE3 on the tissue microarray. It has a predominantly chondroid morphology; however, this tumor was brachyury and S100 protein positive (original magnification 100, insets: × 400). On the original whole-section immunostain for cytokeratin (not shown) it was focally positive. Abbreviation: AE1/3, cytokeratin AE1/AE3.
Single- and multiple-marker performance characteristics for the discrimination between chordoma and chondrosarcoma are summarized in Table 4. Overall, AE1/AE3 had had the best single-marker performance on the tissue microarray with a sensitivity of 97% and a specificity of 100% (accuracy 97%) for the diagnosis of chordoma. Brachyury followed, showing a 90% sensitivity and 100% (accuracy 92%). EMA alone had a sensitivity of 94% with a specificity of 71% (accuracy 88%). CD24, CEA, and GFAP did not perform as well, each with an overall accuracy of less than 60% for the diagnosis of chordoma. Of the chondrosarcoma-selective markers, podoplanin as the sole discriminating stain showed a sensitivity of 78% and a specificity of 86%. SOX-9 showed essentially no discriminatory ability with a sensitivity of 94%, but specificity of 2%. The most accurate combination of markers using the condition of either one being positive was AE1/AE3+brachyury. This combination showed an improved sensitivity of 98% while retaining specificity of 100% with an overall accuracy of 99%.
Discussion
For the purposes of this study, we examined 35 conventional chordomas, 45 chondroid chordomas, and 24 chondrosarcomas of the skull base representing 104 total tumors from 70 patients spanning 38 years. Although our objective was not to reiterate the discriminatory histologic features between chordoma and chondrosarcoma, our opinion is similar to the findings of Rosenberg et al,18 in that with adequate sampling and histologic quality, the distinction between chordoma and chondrosarcoma can be reliably made using morphology alone. However, in our experience, the diagnostic material provided for skull base lesions may often be scant and crushed to the point of essentially mandating confirmatory immunostains. The combination of S100 protein and cytokeratin has been the standard immunohistochemical panel used to discriminate chordoma from chondrosarcoma.12, 23, 27 Again, this simple panel is generally reliable in the distinction between chordoma and chondrosarcoma. However, as noted above, poor tissue quality may also adversely impact immunostaining.41, 42 In addition, fixation protocols for lesions vary between laboratories, and we noted that almost two-thirds of our cases were decalcified.41, 42, 43 Yet another concern includes actual marker heterogeneity within a tumor leading to false negativity.44, 45 Finally, albeit rarely, the differential diagnosis for chordoma (and chondrosarcoma) of the skull base also spans other entities like chordoid meningioma, myoepithelioma, and metastatic clear cell carcinomas.46 For some of these diagnostic considerations, keratin and S100 protein coexpression alone will not separate these lesions. Thus, other markers that support the traditional markers are desirable.
Here, we used the increasingly popular tissue microarray methodology to efficiently assess the ability of the new immunohistochemical markers podoplanin, brachyury, and SOX-9, to better differentiate chordomas and chondrosarcomas. The advantages and disadvantages of tissue microarray are well described in the literature.44, 47, 48 Although this is a tissue conserving high throughput methodology, it has drawbacks that include core loss and tissue heterogeneity. In chondroid chordomas, any potential antigenic heterogeneity is compounded further by the morphologic heterogeneity. Both of these issues are best mitigated (but not completely resolved) by using multiple replicates of cores in the microarray construction targeting specific morphologic areas. We used cores arrayed in triplicate in the present study and attempted to target both components of chondroid chordomas. However, our series showed a case loss rate of 25–29% per stain, which is much higher than reported16, 45, 49, 50 and in our own experience with non-chondroid lesions (0–5%; data not published). It is likely that the hardness of many of the tissues, combined with the epitope retrieval methods used, contributed to this degree of core loss.51
Our results show that cytokeratin staining retains the best individual performance in differentiating chordoma from chondrosarcoma. Only two chordomas (both chondroid) were keratin negative on the tissue microarray. Both of these did, however, show patchy keratin reactivity on the corresponding original diagnostic material suggesting that tumor heterogeneity may have affected all the immunostains that we had tested. Although tumoral heterogeneity is generally considered a limitation of tissue microarray methodology, this limitation does simulate, albeit in an extreme fashion, the problems encountered when assessing small diagnostic biopsies. As expected, because of its function driving mesoderm and notochord differentiation, staining for the transcription factor brachyury was entirely specific for chordoma. However, brachyury was not as sensitive as cytokeratin. Potential reasons for this may include again, tumoral heterogeneity, fixation, and/or decalcification. However, as half of the brachyury-negative chordomas showed solid poorly differentiated areas, brachyury loss may represent a form of tumor progression. Indeed, there was a trend toward a poor outcome in brachyury-negative chordomas. A decrease in reactivity has been noted in sarcomatoid component in a dedifferentiated chordoma reported previously.16 Combining brachyury and cytokeratin AE1/AE3 offered the best accuracy, demonstrating the advantage, though slight, of a panel-based approach over a single-marker approach. In addition, although not specifically investigated in this study, brachyury would help to differentiate chordoma from other cytokeratin-positive soft tissue lesions of the skull base, such as myoepithelioma and metastatic clear cell carcinoma, as suggested by Tirabosco et al,46 in their study of extra-axial chordomas in comparison to soft tissue myoepitheliomas. Negative staining for brachyury and cytokeratin AE1/AE3 was almost equally as powerful an identifier of chondrosarcoma. EMA was also highly sensitive for chordomas, but not entirely specific, staining a quarter of chondrosarcomas as well. EMA reactivity in chondrosarcoma has been noted in up to 8% of skull base chondrosarcomas in one large series of 200 cases. Chondrocytes in myxoid areas of chondrosarcoma have been noted to be occasionally EMA positive.52 Our percentage was rather high, but this may be an artifact of our lower number of cases (16 for this stain).
Regarding positive markers of chondrosarcoma, podoplanin did not perform as reported previously by Huse et al.28 Although podoplanin was positive in the great majority of our chondrosarcomas (78%), this is somewhat fewer than the 94% reported previously.28 In addition, our study did show podoplanin positivity in 15% of chordomas. Our larger sample size, particularly regarding inclusion of more chondroid chordomas, is the likely explanation. Considering that our podoplanin-positive chordomas were mostly chondroid, podoplanin is likely a marker of a well-developed chondroid phenotype rather than chondroid histogenesis. In addition, podoplanin results underperformed cytokeratin and brachyury individually, and did not add accuracy to any of the top combinations of markers. In contrast, the presumed chondroid lineage marker SOX-9 showed virtually no discriminatory capacity in this particular differential diagnosis. Previous studies in mouse models have described SOX-9 function in the notochord and during chondrogenesis.36, 53 Our results show SOX-9 expression in human tumors of notochord and chondroid differentiation in indirect support of similar functions for this molecule in humans.
CD24, though entirely specific, was not very sensitive in identifying chordoma. This finding was unexpected as Fujita et al33 had previously suggested that CD24 reliably stained seven chordomas, with positive staining for this marker not observed in chondrosarcoma. In our hands, fewer than half of our chordomas were positive. Potential explanations include tissue fixation and decalcification, and our examination of a larger cohort. GFAP and polyclonal CEA did not perform very well in the chordoma/chondrosarcoma differential as suggested previously,30, 31 but may be useful in the diagnosis of other lesions that are occasionally included in the differential diagnosis.
In summary, cytokeratin is still the most accurate ancillary stain in discriminating chordoma from chondrosarcoma, though the addition of brachyury in a panel-based approach does slightly improve accuracy and serves as a useful confirmatory marker. Interestingly, brachyury may be lost in more histologically and clinically aggressive chordomas. However, podoplanin did not perform as previously reported, and the other newer markers such as SOX-9 and CD24 were of minimal utility altogether.
References
Barnes L, Eveson JW, Reichart PA, et al. Pathology and Genetics of Head and Neck Tumours. IARC Press: Lyon, 2005.
Dorfman HD, Czerniak B . Bone tumors. Mosby: St. Louis, MO, 1998.
Unni KK . Tumors of the Bones and Joints. American Registry of Pathology in collaboration with the Armed Forces Institute of Pathology: Washington, DC, 2005.
Barnes L . Surgical Pathology of the Head and Neck 2nd edn Dekker: New York, 2000.
Heffelfinger MJ, Dahlin DC, MacCarty CS, et al. Chordomas and cartilaginous tumors at the skull base. Cancer 1973;32:410–420.
Rassekh CH, Nuss DW, Kapadia SB, et al. Chondrosarcoma of the nasal septum: skull base imaging and clinicopathologic correlation. Otolaryngol Head Neck Surg 1996;115:29–37.
Sen CN, Sekhar LN, Schramm VL, et al. Chordoma and chondrosarcoma of the cranial base: an 8-year experience. Neurosurgery 1989;25:931–940; discussion 940–931.
Brooks JJ, LiVolsi VA, Trojanowski JQ . Does chondroid chordoma exist? Acta Neuropathol 1987;72:229–235.
Brooks JJ, Trojanowski JQ, LiVolsi VA . Chondroid chordoma: a low-grade chondrosarcoma and its differential diagnosis. Curr Top Pathol 1989;80:165–181.
Ishida T, Dorfman HD . Chondroid chordoma versus low-grade chondrosarcoma of the base of the skull: can immunohistochemistry resolve the controversy? J Neurooncol 1994;18:199–206.
Jeffrey PB, Biava CG, Davis RL . Chondroid chordoma. A hyalinized chordoma without cartilaginous differentiation. Am J Clin Pathol 1995;103:271–279.
Rosenberg AE, Brown GA, Bhan AK, et al. Chondroid chordoma—a variant of chordoma. A morphologic and immunohistochemical study. Am J Clin Pathol 1994;101:36–41.
Mitchell A, Scheithauer BW, Unni KK, et al. Chordoma and chondroid neoplasms of the spheno-occiput. An immunohistochemical study of 41 cases with prognostic and nosologic implications. Cancer 1993;72:2943–2949.
Almefty K, Pravdenkova S, Colli BO, et al. Chordoma and chondrosarcoma: similar, but quite different, skull base tumors. Cancer 2007;110:2457–2467.
Romeo S, Hogendoorn PC . Brachyury and chordoma: the chondroid-chordoid dilemma resolved? J Pathol 2006;209:143–146.
Vujovic S, Henderson S, Presneau N, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol 2006;209:157–165.
Gay E, Sekhar LN, Rubinstein E, et al. Chordomas and chondrosarcomas of the cranial base: results and follow-up of 60 patients. Neurosurgery 1995;36:887–896; discussion 896–887.
Rosenberg AE, Nielsen GP, Keel SB, et al. Chondrosarcoma of the base of the skull: a clinicopathologic study of 200 cases with emphasis on its distinction from chordoma. Am J Surg Pathol 1999;23:1370–1378.
Martin JJ, Niranjan A, Kondziolka D, et al. Radiosurgery for chordomas and chondrosarcomas of the skull base. J Neurosurg 2007;107:758–764.
Cho YH, Kim JH, Khang SK, et al. Chordomas and chondrosarcomas of the skull base: comparative analysis of clinical results in 30 patients. Neurosurg Rev 2008;31:35–43; discussion 43.
Juliao SF, Rand N, Schwartz HS . Galectin-3: a biologic marker and diagnostic aid for chordoma. Clin Orthop Relat Res 2002;70–75.
Mori K, Chano T, Kushima R, et al. Expression of E-cadherin in chordomas: diagnostic marker and possible role of tumor cell affinity. Virchows Arch 2002;440:123–127.
Miettinen M . Chordoma. Antibodies to epithelial membrane antigen and carcinoembryonic antigen in differential diagnosis. Arch Pathol Lab Med 1984;108:891–892.
Miettinen M, Lehto VP, Dahl D, et al. Differential diagnosis of chordoma, chondroid, and ependymal tumors as aided by anti-intermediate filament antibodies. Am J Pathol 1983;112:160–169.
Abenoza P, Sibley RK . Chordoma: an immunohistologic study. Hum Pathol 1986;17:744–747.
Coffin CM, Swanson PE, Wick MR, et al. An immunohistochemical comparison of chordoma with renal cell carcinoma, colorectal adenocarcinoma, and myxopapillary ependymoma: a potential diagnostic dilemma in the diminutive biopsy. Mod Pathol 1993;6:531–538.
Meis JM, Giraldo AA . Chordoma. An immunohistochemical study of 20 cases. Arch Pathol Lab Med 1988;112:553–556.
Huse JT, Pasha TL, Zhang PJ . D2-40 functions as an effective chondroid marker distinguishing true chondroid tumors from chordoma. Acta Neuropathol 2007;113:87–94.
Walker WP, Landas SK, Bromley CM, et al. Immunohistochemical distinction of classic and chondroid chordomas. Mod Pathol 1991;4:661–666.
Wojno KJ, Hruban RH, Garin-Chesa P, et al. Chondroid chordomas and low-grade chondrosarcomas of the craniospinal axis. An immunohistochemical analysis of 17 cases. Am J Surg Pathol 1992;16:1144–1152.
Wittchow R, Landas SK . Glial fibrillary acidic protein expression in pleomorphic adenoma, chordoma, and astrocytoma. A comparison of three antibodies. Arch Path Lab Med 1991;115:1030–1033.
Henderson SR, Guiliano D, Presneau N, et al. A molecular map of mesenchymal tumors. Genome Biol 2005;6:R76.
Fujita N, Miyamoto T, Imai J, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun 2005;338:1890–1896.
Cleaver O, Krieg PA . VEGF mediates angioblast migration during development of the dorsal aorta in Xenopus. Development 1998;125:3905–3914.
Ng LJ, Wheatley S, Muscat GE, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol 1997;183:108–121.
Lefebvre V, Huang W, Harley VR, et al. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol 1997;17:2336–2346.
Soderstrom M, Bohling T, Ekfors T, et al. Molecular profiling of human chondrosarcomas for matrix production and cancer markers. Int J Cancer 2002;100:144–151.
Wehrli BM, Huang W, De Crombrugghe B, et al. Sox9, a master regulator of chondrogenesis, distinguishes mesenchymal chondrosarcoma from other small blue round cell tumors. Hum Pathol 2003;34:263–269.
Kordes U, Hagel C . Expression of SOX9 and SOX10 in central neuroepithelial tumor. J Neurooncol 2006;80:151–155.
Zhao C, Bratthauer GL, Barner R, et al. Immunohistochemical analysis of sox9 in ovarian Sertoli cell tumors and other tumors in the differential diagnosis. Int J Gynecol Pathol 2007;26:1–9.
Goldstein NS, Hewitt SM, Taylor CR, et al. Recommendations for improved standardization of immunohistochemistry. Appl Immunohistochem Mol Morphol 2007;15:124–133.
Taylor CR . The total test approach to standardization of immunohistochemistry. Arch Pathol Lab Med 2000;124:945–951.
O'Leary TJ . Standardization in immunohistochemistry. Appl Immunohistochem Mol Morphol 2001;9:3–8.
Linderoth J, Ehinger M, Akerman M, et al. Tissue microarray is inappropriate for analysis of BCL6 expression in diffuse large B-cell lymphoma. Eur J Haematol 2007;79:146–149.
Fasig JH, Dupont WD, Olson SJ, et al. Steroid hormone receptor and COX-2 expression in chordoma. Am J Clin Pathol 2007;128:375–381.
Tirabosco R, Mangham DC, Rosenberg AE, et al. Brachyury expression in extra-axial skeletal and soft tissue chordomas: a marker that distinguishes chordoma from mixed tumor/myoepithelioma/parachordoma in soft tissue. Am J Surg Pathol 2008;32:572–580.
Takikita M, Chung JY, Hewitt SM . Tissue microarrays enabling high-throughput molecular pathology. Curr Opin Biotechnol 2007;18:318–325.
Pick E, McCarthy MM, Kluger HM . Assessing expression of apoptotic markers using large cohort tissue microarrays. Methods Mol Biol 2008;414:83–93.
Au NH, Gown AM, Cheang M, et al. P63 expression in lung carcinoma: a tissue microarray study of 408 cases. Appl Immunohistochem Mol Morphol 2004;12:240–247.
Morrison C, Merati K, Marsh Jr WL, et al. The mucin expression profile of endometrial carcinoma and correlation with clinical-pathologic parameters. Appl Immunohistochem Mol Morphol 2007;15:426–431.
Shi SR, Key ME, Kalra KL . Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem 1991;39:741–748.
Weiss SW, Goldblum JR . Enzinger and Weiss's Soft Tissue Tumors. 5th edn Mosby: St. Louis, MO, 2008.
Barrionuevo F, Taketo MM, Scherer G, et al. Sox9 is required for notochord maintenance in mice. Dev Biol 2006;295:128–140.
Acknowledgements
This work was supported by a pilot from the Head and Neck SPORE NIH 1P50 CA097190 (awarded to RRS). The study was also supported by the Stout Family Fund for Head and Neck Cancer Research at the Eye & Ear Foundation of Pittsburgh and the Head and Neck Oncology Registry. We thank Jennifer Ridge-Hetrick and Kim Marie Adams for their administrative support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors have no financial disclosures pertaining to this research to report.
Rights and permissions
About this article
Cite this article
Oakley, G., Fuhrer, K. & Seethala, R. Brachyury, SOX-9, and podoplanin, new markers in the skull base chordoma vs chondrosarcoma differential: a tissue microarray-based comparative analysis. Mod Pathol 21, 1461–1469 (2008). https://doi.org/10.1038/modpathol.2008.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.144
Keywords
This article is cited by
-
Organelle resolved proteomics uncovers PLA2R1 as a novel cell surface marker required for chordoma growth
Acta Neuropathologica Communications (2024)
-
Virus-like particle-based delivery of Cas9/guide RNA ribonucleoprotein efficiently edits the brachyury gene and inhibits chordoma growth in vivo
Discover Oncology (2023)
-
Chordoma: demographics and survival analysis with a focus on racial disparities and the role of surgery, a U.S. population-based study
Clinical and Translational Oncology (2023)
-
LIM and SH3 protein 1 (LASP1) differentiates malignant chordomas from less malignant chondrosarcomas
Journal of Neuro-Oncology (2022)
-
Primäres extraaxiales chondroides Chordom des anterioren Nasenseptums: Fallbericht eines seltenen Chordoms mit Literaturübersicht
HNO (2021)