Abstract
The intestinal epithelial cells constitute the first line of defense against gut microbes, which includes secretion of various antimicrobial substances. Reactive oxygen species (ROS) are well characterized as part of the innate phagocytic immunity; however, a role in controlling microorganisms in the gut lumen is less clear. Here, we show a role for nitric oxide synthase (iNOS)- and NOX1-produced ROS in maintaining homeostasis of the gut microbiota. In vivo imaging revealed distinctly high levels of ROS in the ileum of normal healthy mice, regulated in accordance with the amount of gut bacteria. The ROS level was dependent on the nitric oxide and superoxide producers iNOS and NOX1, respectively, suggesting peroxynitrite as the effector molecule. In the ileum of iNOS- and NOX1-deficient mice, the bacterial load is increased and the composition is more cecum like. Our data suggest a unique role of ileum in maintaining homeostasis of gut microbes through production of ROS with potential importance for preventing reflux from the large intestine, bacterial overgrowth, and translocation.
Similar content being viewed by others
Introduction
The small intestine possesses a dual role: on the one hand, it is responsible for uptake of nutrients, which requires a large absorptive surface, whereas on the other hand it must prevent the gut lumen microbiota from entering this same surface. The gut lumen is separated from the host interior by only one cell layer, which is ideal for its absorptive role. However, this creates a challenge for its role as a barrier. As part of the barrier function, the epithelial cells interact with the microbes in the gut lumen. Several secretory molecules, such as mucins that make up the mucus layer, antimicrobial peptides, and secretory IgA participate in controlling the gut microbes. Reactive oxygen species (ROS) are universally accepted to be an essential component of the innate immune system through the respiratory burst in neutrophils, macrophages, and dendritic cells; however, an antimicrobial role of ROS in the gut is less determined. Here, we have explored a role of ROS production in maintaining bacterial homeostasis in the small intestine at the border with the large intestine.
The ROS producing enzymes, Dual oxidase (DUOX) and NADPH oxidase (NOX), are expressed in the intestinal epithelial cells in correlation with microbial content, indicating a role of these enzymes in ROS-based immune homeostasis of the gut.1 In Drosophila, it is shown that DUOX produces ROS in the gut, which is directly bactericidal, and necessary to control gut bacteria and prevent infections. The DUOX in Drosophila contains two enzymatic domains: one oxidase domain that produces hydrogen peroxide and one peroxidase domain that converts the hydrogen peroxide to hypochlorous acid.2 In C. elegans, DUOX-dependent ROS production in the gut was shown to improve survival during bacterial infections; however, no direct bactericidal effect was determined.3 Hydrogen peroxide is suggested as the effector molecule, but this is a less bactericidal molecule than hypochlorous acid. In mammals, DUOX2 is reported to have an anti-bacterial role in cultured primary airway epithelial cells4 and in defense against Helicobacter felis infection in gastric epithelium.5 In these systems, it was suggested that DUOX2 produced hydrogen peroxide as a substrate for lactoperoxidase, with a subsequent formation of the bactericidal hypothiocyanous acid. In a DUOX2-deficient mouse, bacterial translocation was increased and expression of a ROS-inducible gene was downregulated in the mucosal-segmented filamentous bacteria.6 NOX1 is highly expressed in epithelial cells, particularly, in colon and ileum. Most studies have focused on NOX1’s role in redox modifications of signaling pathways involved in cell division and differentiation, particularly, with respect to wound healing in colon.7 However, its role in controlling gut microbes is elusive. In one study of NOX1 knockout (KO) mice inoculated with Salmonella typhimurium, no effect on cecal concentration of the bacterium and changes in overall protection were observed.8 In another study with Citrobacter rodentium infection, reduced cecal numbers and lower disease status were found in NOX1 KO mice.9 They further provided evidence suggesting NOX1 to operate through regulation of DUOX2. The NOX enzymes generate superoxide, which does not kill microbes directly, but is precursor of other ROS with bactericidal activity such as hydrogen peroxide, hydroxyl radicals, hypochlorous acid, and peroxynitrite. Superoxide combines with nitric oxide (NO) to form peroxynitrite, which is regarded as one of the crucial bactericidal ROS in the respiratory burst of the innate immune system. NO is produced by inducible nitric oxide synthase (iNOS), which is expressed by epithelial cells of the small intestine in association with the amount of bacterial content.1, 10 Furthermore, NO is found in the gut lumen.11 This raises the question whether peroxynitrite produced in the intestine can affect the luminal gut bacteria and thus participate in maintaining homeostasis of gut microbes.
Here, we have found evidence for a high NOX1- and iNOS-dependent ROS production in the ileum of normal healthy mice. We furthermore observed a strong impact of this ROS production on the microbial load and composition in the ileum. This suggests a unique role of ileum in maintaining homeostasis of gut microbes through production of peroxynitrite with potential importance for preventing reflux from the large intestine, bacterial overgrowth, and invasion.
Results
iNOS- and NOX1-dependent production of ileal ROS
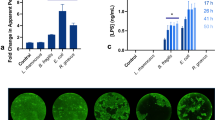
To determine whether ROS are present in the small intestine, we used the optical imaging probe L-012, which has high specificity for ROS, particularly, peroxynitrite.12 In vivo imaging of L-012-dependent luminescence showed a strong signal coming from the abdominal region of the mice (Figure 1a). Dissection and imaging of individual organs ex vivo showed that the signal originated from the distal small intestine, the ileum (Figure 1b). This overlaps with the particularly high expression of iNOS in ileum in comparison to the rest of the intestine and the relatively high expression of NOX1 in ileum compared to more proximal parts (Figure 1c). As these enzymes catalyze the production of superoxide anion and NO, which rapidly react to form peroxynitrite we wanted to determine their involvement in the ROS production. We injected the NOS inhibitor L-NAME i.v. (50 mg kg-1) and imaged the L-012-mediated luminescence, which was reduced by 70% (Figure 1d). To elucidate the involvement of superoxide, we added the superoxide scavenger TEMPOL in the drinking water (2.5 g l-1) (Figure 1e). After 2 days, the L-012 signal was reduced by more than 60%. To assess the presence of peroxynitrite, we administrated the peroxynitrite scavenger MnTBAP chloride i.v (20 mg kg-1), and we here observed around 60% reduction in the L-012 signal (Figure 1f). Furthermore, we explored the ROS production in KO mice of iNOS and NOX1. We first imaged the L-012 signal in KO mice cohoused with wild type (WT) mice and then 4 weeks after being separated and housed as single genotypes (Figure 1g). The signal was reduced by around 60% in the iNOS KO mice and almost abolished in the NOX1 KO mice. We did not observe any significant difference between the mice during and after cohousing. Furthermore, as peroxynitrite is a strong oxidant, we gave the antioxidants vitamin C (10 g l-1) and tannic acid (5 g l-1) in the drinking water of WT mice. We measured the L-012 signal over 3 weeks (Figure 1h). The signal went gradually down reaching 70% reduction for both antioxidants. To determine whether the epithelial cells could be responsible for the peroxynitrite production, we measured the NO content in these cells with the probe DAF-FM using flow cytometry (Figure 2a, b). We observed in the cells staining positively for the epithelial cell marker CD326, a significantly larger fraction of cells with DAF-FM labeling in the distal small intestine compared with the proximal region. Furthermore, the distal cells showed higher mean fluorescent intensity indicating more NO production in the distal cells compared to the proximal. As innate immune cells in the lamina propria can mount a respiratory burst, we also determined the NO content in cells stained with CD11b, a marker for monocytes, dendritic cells, and macrophages. There was significantly higher signal from the epithelial cells in comparison to CD11b stained cells. Taken together, these observations support an iNOS- and NOX1-dependent ROS production in epithelial cells of ileum.
iNOS- and NOX1-dependent production of ileal ROS. (a, b) L-012-mediated luminescence signal (a) in vivo and (b) ex vivo indicating high ROS production in the terminal part of the small intestine, the ileum. Dissected organs displayed to the right in the panel illustrates low L-012 signal (from top to bottom: skin, liver, pancreas, spleen, inguinal lymph node, kidney and mesenteric fat). The pseudo colors represent photons s-1 cm2 sr. (c) mRNA expression of iNOS and NOX1 in the small intestine and colon of WT mice. GADPH is used for normalization. Both of the genes are highly expressed in ileum, n=5. (d–h) The following conditions reduced the L-012-mediated signal: (d) L-NAME provided i.p., n=6; (e) TEMPOL in the drinking water, n=8–10; (f) MnTBAP provided i.v., n=8 (g) iNOS and NOX1 KO mice compared to WT that were imaged both during cohousing with WT mice and 4 weeks after separation to individual cages, n=15–30; (h) Vitamin C or tannic acid in the drinking water, n=15. Values are mean with s.e.m. (c) Student’s t-test. (d–f) Student’s t-test of the difference in reduction between control and treatment groups. (g) Kruskal–Wallis test with Dunn’s correction for multiple comparisons. (h) One-way ANOVA. iNOS, nitric oxide synthase; ROS, reactive oxygen species; WT, wild type.
Flow cytometry analysis of NO content in cells of proximal and distal part of the small intestine. (a) Scatter plots of the NO probe DAF-FM staining combined with the epithelial cell marker CD326 and the phagocyte cell marker CD11b in cells isolated from the epithelial layer and lamina propria. Labeling of cells from iNOS KO is consistent with the absence of NO production (b) WT and iNOS KO mice show higher NO production in epithelial cells of distal intestine compared to both proximal intestine and macrophages of the lamina propria Mann–Whitney test. Furthermore, in all the corresponding cell populations of the iNOS KO and WT mice, the iNOS KOs showed significantly reduced NO production (P<0.03 for all comparisons) Kruskal–Wallis test with Dunn’s correction for multiple comparisons. Values are mean with s.e.m. iNOS, nitric oxide synthase; NO, nitric oxide; WT, wild type.
The amount of gut bacteria is associated with the ROS production
To elucidate whether gut bacteria can induce ROS production in ileum, we manipulated the amount of bacteria. To decrease the bacterial load, we first used an antibiotics treatment proven to almost fully deplete the gut microbiota (1 g l-1 neomycin, 0.5 g l-1 vancomycin, 1 g l-1 metronidazole, and 1 g l-1 ampicillin, oral gavage daily for a week).13 As this is a harsh treatment with potential unforeseen physiological effects and depletion of ileal microbiota is sufficient here, we also exposed the mice to a milder antibiotic treatment (0.5 g l-1 neomycin and 0.25 g l-1 vancomycin through the drinking water over 4 weeks) (Figure 3a, b). In both cases, the L-012-mediated signal went down by ∼80%. As expected, the bacterial load went down in ileum (Figure 3c). Consistent with these observations, the mRNA expression of both iNOS and NOX1 was decreased in the ileum (Figure 3d, e). We also decreased the bacterial content in the intestine by 15 h fasting (Figure 3f). This caused a reduction in the L-012 signal from the mice by 70%. To study the ROS production during an increase in the bacterial load, we imaged mice before, during and after weaning, as this is a period where massive increase of intestinal bacteria occurs.14 Here, we observed a large increase in the L-012 signal the first week after separating the mice from their mother, which thereafter stabilized (Figure 3g). In conclusion, these finding suggest that the intestinal ROS production is dependent on the presence of bacteria in the gut.
Effect of bacterial luminal load on ROS production. (a, b) The L-012-mediated signal is reduced in mice receiving antibiotics concoctions either by (a) daily oral gavage, n=5–6 or (b) via the drinking water, n=10. (c) The amount of cultivable luminal chyme bacteria in ileum is reduced in mice receiving antibiotics via gavage over 1 week, n=4–5. (d, e) The mRNA expression of iNOS and NOX1 decreases in jejunum and ileum in mice receiving antibiotics in the drinking water over 4 weeks, n=10. (f) The L-012-mediated signal goes down in mice after 15 h of fasting n=10. (g) The L-012 signal is minimal in the 3 weeks prior to weaning and increases severely after separation from mother n=11–16. Values are mean with s.e.m. (a, f) Student’s t-test of the difference in reduction between control and treatment groups. (c) Student’s t-test (b, g) one-way ANOVA. (d, e) One-way ANOVA with Sidak’s correction for multiple comparisons. iNOS, nitric oxide synthase; ROS, reactive oxygen species.
iNOS and NOX1 KO mice had higher bacterial load in the distal small intestine
As we observed that iNOS and NOX1 are necessary for the ROS production, we sought to determine whether the bacterial content in the gut of iNOS and NOX1 KO mice is increased compared to WT mice. To balance the environmental influence, the KO and WT mice were cohoused for 3 to 4 weeks directly after weaning. The mice were then allocated to separate cages based on genotype for 4 weeks before sampling. We estimated the amount of bacteria in the luminal chyme by colony-forming units (CFU) counting (Figure 4a) and in the mucus by RT-qPCR against the 16S rRNA gene (Figure 4b).). In both KO types, the amount of bacteria was higher in ileum and jejunum in comparison to the WT mice for the luminal samples. For the mucosal samples, we did observe higher bacterial load in the ileum of the two KOs, but it was only statistically significant for the NOX1. We also estimated the bacterial content in cecum samples by CFU counting (Figure 4a). Here, we found no significant difference between either of the KO with the WT mice. Interestingly, we observed that in the KOs the bacterial amount reached cecum-like numbers in ileum, consistent with potential bacterial overgrowth. In conclusion, the amount of bacteria in the distal small intestine is dependent on both iNOS and NOX1 suggesting involvement of a converging effect, possibly the formation of peroxynitrite.
ROS production in the distal part of the small intestine affects the amount of mucosal and luminal bacteria. (a) Amount of cultivable bacteria in the luminal chyme of iNOS and NOX1 KO mice. In both jejunum and ileum, the amount of bacteria is increased in the KO mice in comparison to their, respectively, cohoused WT mice. In cecum, no such difference could be detected, n=10–16. (b) Amount of bacteria in the mucus layer as determined by RT-qPCR of 16S rRNA. GADPH is used for normalization. The bacterial load is increased in the jejunum and ileum of the NOX1 KO mice, n=9–15. Values are mean with s.e.m. (a) One-way ANOVA with Sidak’s correction for multiple comparisons. (b) Kruskal–Wallis test with Dunn’s correction for multiple comparisons. iNOS, nitric oxide synthase; ROS, reactive oxygen species; WT, wild type.
The microbial composition in the small intestine is altered in a similar manner for the iNOS and NOX1 KO mice
To determine the impact of ROS production on potential alterations in the composition of the gut microbiota, we performed 16S rRNA gene sequencing of DNA extracted from mucosa and luminal chyme samples of the WTs and the iNOS and NOX1 KO mice. We observed that at phylum level, there is a similar shift in the microbial composition in jejunum and ileum for both KO mice (Figure 5a). Firmicutes increase and Bacteroidetes concurrently decrease. This can be seen for both mucosa and luminal chyme samples; however, it is more pronounced in the former. In cecum, the microbial composition appears to be unaltered between the KO and the WT mice. To investigate whether the gut microbiota is also altered at finer taxonomic levels, we performed a Principal component analysis (PCA) using the relative abundance of the taxa classified at family level (Figure 5b–e). Families that were present in at least five mice with relative abundance higher than 0.5% were included in the analysis. The PCA distinguished the ileal samples of the WT and KO, whereas the two KO types co-clustered. The results were similar for the mucus and chyme samples; however, the chyme samples show some degree of overlap with the WT. All cecum samples showed co-clustering. In addition, we sought to determine whether there is an overlap between the specifically differentiated taxa for each of the two KO mice. We performed a LEfSE analysis on the ileal mucosal samples to identify the taxa that were associated with the iNOS and the NOX1 KO mice. Five taxa were identified for the iNOS KO mice: four increasing (Actinobacteria, Actinomycetales, Sporosarcina, Allobaculum) and one decreasing (S24_7). For the NOX1 KO mice, nine taxa were identified: eight increasing (Actinobacteria, Bacillaceae, Coriobacteriaceae, Dietzia, Turicibacter, Bifidobacterium, Sporosarcina, Allobaculum) and one decreasing (Lactobacillus). We next compared the relative abundance of the enriched identified taxa in each of the KOs to the WT (Figure 6). Six out of the eight taxa associated with the NOX1 KO mice show a statistically significant increase in the iNOS KO mice, whereas three out of the four taxa associated with the iNOS KO mice increase in the NOX1 KO mice. However, the taxa that decreased in each of the KOs did not change significantly in the other KO type. Taken together, these findings suggest that iNOS- and NOX1-dependent production of ROS in the small intestine alters the gut microbial composition. The observed alterations appear to be similar for both KOs indicating a synergistic effect of NO and superoxide.
Gut microbial composition of the iNOS and NOX1 KO mice shows overlap. (a) Relative abundance of phyla in iNOS and NOX1 KO mice and their, respectively, cohoused WT mice. In the two KO mice, Firmicutes are increased in the mucus layer of both jejunum (P<0.0001 for both KO) and ileum (P<0.0001 for iNOS KO and P=0.01 for NOX1 KO), whereas Bacteroidetes decrease (jejunum P<0.0001 for both KO, ileum P<0.0001 for iNOS KO and P=0.006 for NOX1 KO). For the ileal chyme samples, Firmicutes are statistically increased only in NOX1 KO (P=0.004), whereas Bacteroidetes increase only in jejunum in iNOS KO (P=0.02) and in ileum in NOX1 KO (P=0.0002). No differences could be detected in the cecum samples, n=10–16. Values are mean with s.e.m. One-way ANOVA with Tukey’s post hoc. (b, c) PCA plot of the taxa at family level of the ileum and cecum samples. After examining all possible principal components, we found that PC-1 and PC-3 could best explain the variation among the groups. All of the cecum samples were co-clustered regardless of mouse groups. (b) The ileal mucosa samples of the two KO mice cluster together and the two WT mice cluster together with minimal overlap between the KOs and the WTs, n=8–15. For the ileal chyme samples (c) a similar pattern is observed; however, more overlap can be seen between the iNOS and the WT, n=10–15. (d, e) Loading scores of cecum, ileal mucosa (d), and ileal chyme (e) samples presented in PCA plots in b and c. In accordance with the above findings, both KO mice for both of the ileal compartments differ from their respective WT on PC3, whereas the iNOS mucosal samples are also separated in PC1, n=8–15. Values are mean with s.e.m. One-way ANOVA with Sidak’s corrections for multiple comparisons. iNOS, nitric oxide synthase; PCA, principal component analysis; ROS, reactive oxygen species; WT, wild type.
Overlap in identified taxa between iNOS and NOX1 KO mice. Relative abundance of the taxa that either increased or decreased in each of the KO mice as identified by LEfSE. The two KO types show a similar increase for most of these taxa, n=10–14. Values are mean with s.e.m. For each taxon after logarithmic data transformation, one-way ANOVA with Sidak’s correction for multiple comparisons. iNOS, nitric oxide synthase; LEfSE, linear discriminant analysis effect size.
iNOS and NOX1 KO mice show a shift in ileal microbiota resembling more cecal composition
We observed a higher ratio of Firmicutes to Bacteroidetes for the two KOs (Figure 7a). Interestingly, this moves the microbiota composition in a direction resembling more the one in cecum. Furthermore, the increased Firmicutes/Bacteroidetes ratio is most pronounced in the ileum and becomes gradually less in more proximal parts of the small intestine. To elucidate whether this could be caused by migration of bacteria from cecum to ileum, we identified all genera that were significantly more abundant in cecum than in ileum of WT mice and determined if they were increased in the KO mice (Figure 7b). We observed the abundance in ileum of most of these genera to be higher in both of the two KO mouse types compared to WT. In conclusion, these results may imply that ROS production in ileum has a role in prevention of cecal microbiota reflux.
The gut microbial composition of the iNOS and NOX1 KO mice shifts in the direction of cecum. (a) Ratio of relative abundance of Firmicutes and Bacteroidetes in different mucosa gut segments. The ratio increases for both KO mice in jejunum and in ileum. No difference was observed for duodenum, n=11–16. Values are mean with s.e.m. One-way ANOVA with Sidak’s correction for multiple comparisons for jejunum and duodenum samples, Kruskal–Wallis test with Dunn’s correction for multiple comparisons for ileum samples. (b) Shift in the relative abundance of the eight genera that were higher in cecum compared to the small intestine. The heat map shows the ratio of ileum to cecum. Columns represent the mean value of each group. In general, in both KOs the mice showed warmer colors compared to the WTs, indicating an ileal shift in these genera to a cecum-like profile, n=8–12. One-way ANOVA. iNOS, nitric oxide synthase; WT, wild type.
Bacterial DNA is higher in the liver of the KO mice
To evaluate potential implications of the intestinal ROS production on bacterial translocation, we determined the content of bacterial DNA in liver by RT-qPCR against the 16S rRNA gene. We found an increase in bacterial DNA of 56% (n=6–10, P=0.02, Kruskal–Wallis test with Dunn’s correction for multiple comparisons) for iNOS KO mice and of 112% (n=10–13, P=0.009) for NOX1 KO mice compared to WT mice, indicating a role for intestinal ROS production in defense against bacterial translocation.
Discussion
In the present study, we have demonstrated that ROS, possibly peroxynitrite, are highly produced by iNOS and NOX1 in the ileum of normal healthy mice. Furthermore, the amount, composition and translocation of intestinal bacteria appear to be regulated by this ROS production.
To address temporal and spatial in vivo ROS production, we exploited the luminescent probe L-012 as an imaging marker for ROS.15, 16, 17, 18 We observed a strong L-012-mediated signal in ileum that we believe is the result of epithelial cell secretion of superoxide and NO, with a resultant peroxynitrite formation. This is supported by the following: firstly, L-012 is recognized to act extracellularly, and with a high specificity and sensitivity to peroxynitrite.12 NOX1 together with iNOS are responsible for the signal and the major substrate of superoxide, in terms of reaction kinetics, is NO, which together form peroxynitrite.19 Both L-NAME, a NOS inhibitor, and using iNOS KO mice reduced the signal, whereas the superoxide quencher TEMPOL and particularly knocking out NOX1 nearly abolished the L-012-generated signal. Secondly, NOX1 and iNOS are co-expressed in the small intestine, with a particularly high expression in the ileum. In addition, the L-012 signal is not present in the colon where NOX1 has high expression, but where iNOS is not detectable. Finally, we demonstrated that NO production is more abundant in ileal epithelial cells compared to more proximal intestinal regions and immune cells of the lamina propria. Others have reported that iNOS expression in the healthy gut is confined to the ileal villi.10 We regard these data as convincing evidence of NO and superoxide production by epithelial cells of ileum, which may join to form peroxynitrite in the gut lumen.
Although peroxynitrite is likely detected in our experiments, there are other candidates such as hydrogen peroxide and hypochlorous acid, which both have prominent roles in respiratory burst.9 However, hydrogen peroxide is in itself a poor activator of L-012 and requires the presence of high concentrations of peroxidase,20 which to our knowledge is not expressed at high levels in the ileum of normal healthy mice. Hypochlorous acid can activate L-012, but its production is dependent on myeloperoxidase.21 Hydrogen peroxide can also be converted to hydroxyl radicals in the presence of Fe2+ through the Fenton reaction. Hydroxyl radicals can activate L-012, but the concentration of Fe2+ in the intestine is low. However, superoxide can through the Haber–Weiss reaction generate Fe2+ suggesting that a combination of NOX1 and DUOX2 could generate part of the intestinal ROS responsible for the L-012 signal. Indeed, when eliminating the iNOS activity not all the signal is abolished. In addition to ROS production related to immune defense, ROS are also produced by the electron transport chain inside the mitochondria and in connection with intracellular signaling, but this is a global phenomenon and comprise only low levels of ROS concentrations, and hence cannot be imaged in vivo.
The strength of the L-012-mediated signal is exceptionally high, which implies that the amount of ROS produced in the ileum is high. This is in accordance with previous studies where L-012 is used to image ROS during various conditions of acute inflammation.15, 17 Interestingly, the signal strength generated by L-012 in models of arthritis and skin inflammation is lower than the signal we observe in a healthy ileum. However, it resembles the amount of signal in models of colonic inflammation.17, 18 This raises the interesting question if a large amount of ROS production only is consistent with secretion in non-host tissue, such as the intestinal lumen. Certainly, constant exposure to an extensive oxidative environment can be detrimental to the host tissue.
We found that in the ileum of iNOS and NOX1 KO mice the bacterial load was increased, suggesting an antimicrobial role for the epithelium-deriving ROS. This could be seen overall in ileum, both in the luminal content and in the mucosa. However, the approach used for the assessment of the bacteria in the luminal content was based on CFU counting, which only assesses the cultivable bacteria, and thus gives an underestimate of the total bacteria that are present. In addition, the microbial composition of both KO mice was altered. The changes in bacterial composition were similar between NOX1 and iNOS KO mice and to some extent more evident in mucus compared to lumen samples, supporting a cooperative effect of superoxide and nitric oxide originating from the epithelium, as discussed above possibly through the secretion of peroxynitrite. The strong reduction of L-012-mediated signal following administration of broad-spectrum antibiotics, concurred with a downregulation of iNOS and NOX1. This observation fits well with comparable data showing that germ-free mice have four- and sevenfold lower expression of NOX1 and iNOS, respectively, than conventional mice.1 Furthermore, intestinal ROS production is lower in germ-free mice, as detected by a broad range ex vivo ROS probe.22 L-012 signals were also influenced by other conditions that change the bacterial load such as after introduction of solid food in weaned mice and during a fasting period.14, 23 It has been known for some time that ROS affect the luminal bacteria directly. It is reported that during an infectious state DUOX-dependent ROS exert antimicrobial properties in the gut of Drosophila.2 In addition, hydrogen peroxide through the activity of DUOX2 is reported to directly influence the virulence of Campylobacter jejuni in an organ culture.24 In a DUOX2 KO mouse, a ROS-inducible gene was lowered in mucosal associated bacteria suggesting a role of hydrogen peroxide against microbes close to the epithelial surface.6 ROS through the action of NOX1 has foremost been associated with intracellular signaling related to tissue-wound repair in the colon.7, 25 Pircalabioru et al. 9 showed reduced DUOX2 expression in a NOX1 KO mouse during infection suggesting a regulatory role of NOX1 in relation to luminal bacteria. They further observed, in accordance with our results, that microbiota in cecum of the KO had the same distribution of the main phyla as in the WT mice. However, they found an increase in certain species of Lactobacilli. It is difficult to compare their data directly with ours as they did not assess the microbiota composition in ileum. Nevertheless, we found that Lactobacilli are reduced in the ileum of NOX1 KO mice. As Lactobacilli are known to harbor a strong defense against ROS,26, 27 we believe that the reduction of ROS rather removes a competitive advantage that can explain the decrease in Lactobacilli. Our data contribute to the knowledge about NOX1 by showing a direct role in controlling the gut microbiota.
The microbiota content in ileum is much higher than in the rest of the small intestine. This is likely caused by microbial reflux from the large load of microbes in cecum and colon. This suggests a unique role for ileum in preventing bacterial overgrowth and maintaining homeostasis of microbes in the small intestine. This is reflected by the higher density of lymphoid tissue in ileum compared to the rest of the gut. We believe that secretion of ROS, potentially peroxynitrite, from the ileal epithelial cells is an integral part of a defense system controlling the small intestinal microbiota and thus important for the nutrient uptake and barrier role of the gut. These insights may have implications for understanding the pathogenic mechanisms that underlie conditions or diseases often found in the ileum such as SIBO, Crohn’s disease, and ileitis.
Methods
Animals
Animal procedures were approved by the Norwegian Animal Research Authority (Mattilsynet). All experiments apart from the experiments with KO mice were performed on NMRI mice of both genders up to 3 months of age from Janvier Labs. In experiments where both genders were used, we could not observe any gender-specific differences. Breeding stocks of C57BL/6J, and the following KO mice: iNOS−/− and NOX1−/−, y/- on C57BL/6J background were purchased from The Jackson Laboratory. As the largest impact of environmental influence on gut microbiota composition occurs during weaning, we cohoused the KO mice for 3–4 weeks together with WT mice directly after separation from mother.14, 28 All mice used for the analysis of the gut microbiota composition were supplied by the same provider (The Jackson Laboratory, Bar Harbor, ME). The diet was RM1 diet (SDS Diet, Essex, UK) and mice were housed in humidity, temperature and by 12 h night/day light cycle controlled environment in individually ventilated cages (Innovive, San Diego, CA).
Imaging
Imaging was done with IVIS Lumina II (Perkin Elmer, Walthamn, MA). The luminescent probe L-012 (Wako Chemical, Neuss, Germany) was dissolved in saline and injected intraperitoneally (i.p.) at 10 mg kg-1. During in vivo imaging the mice were immobilized using isoflurane (2.5–3.5%). Ex vivo imaging on dissected organs was performed within 5 min of L-012 injection. Data acquisition were done with the Living Image software (Perkin Elmer). Light emission from the region of interest was quantified as photons s−1 cm−2 steradian−1.
Flow cytometry
Isolation of an epithelial cell fraction and a lamina propria cell fraction was done as previously described by Goodyear et al.29 Briefly, after killing the mouse, the small intestine was dissected and divided into a proximal and a distal part to be analyzed separately. Peyers patches were removed, the intestine was opened longitudinally and cut into 5 mm long pieces and placed in ice-cold RPMI. The tissue successively went through a three-step process removing mucus, isolating epithelial cells, and isolating lamina propria cells. Mucus was removed by incubation in HBSS/5 mM DTT/2% FBS for 20 min at 37 °C with agitation. Epithelial cells were isolated by three incubation steps each in HBSS/5 mM EDTA/2% FBS for 15 min at 37 °C with agitation. Lamina propria cells were isolated by digestion in HBSS/Liberase (Roche, Basel, Switzerland)/DNAse (Sigma Aldrich, St Louis, MO) for 30 min at 37 °C. The epithelial cell fraction and lamina propria cell fraction were further enriched for/or depleted-off epithelial cells using magnetic EpCam microbeads in accordance with the manufacturer's protocol (Miltenyi Biotech, Bergisch Gladbach, Germany). Cells were pre-blocked in 60% inactivated rat sera and 10 μg ml-1 of anti-CD16/32 (HB2.4) in PBS/2%FBS/2 mM EDTA for 10 min on ice. Cells where washed and stained in PBS/2%FBS/2 mM EDTA for 30 min on ice with 5 μl per test of the following antibodies: CD326 Pacific blue (Biolegend, San Diego, CA), CD11b PE-Cy7 (BD Bioscience, San Jose, CA), CD3-APC (BD Bioscience), and CD24-PE (Biolegend). NO was detected with the florescent probe 4-amino-5-methylamino-2′,7′-difluorofluorescein (DAF-FM) in accordance with the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). The labeled cells were acquired on a LSR II flow cytometer (BD Bioscience). Flow data were analyzed in Cytobank.
Sampling
Biological samples were collected under sterile conditions after killing the mice by neck dislocation. The small intestine was divided in three: duodenum (most proximal 5 cm), jejunum (6 cm around the center), and ileum (most distal 6 cm). Cecum samples were taken directly opposite to the ileocecal valve to collect samples close to the valve that additionally are easy to anatomically identify to avoid sampling error. Luminal chyme for DNA extraction or microbe cultivation was squeezed out. The intestinal fragments were cut longitudinally and mucosal samples for RNA or DNA extraction were scraped off with a glass slide.
Counting of cultivable microbes
Luminal chyme samples were weighed, suspended in 1:10 sterile PBS (Sigma Aldrich) and homogenized by brief vortexing. The suspensions were serially diluted in the range of 10−1 to 10−8 and 100 μl of each dilution factor were plated on 7% freshly prepared horse blood agar plates (Oxoid, Basingstoke, UK). The procedure was performed in duplicates and the plates were incubated at 37 °C under anaerobic conditions for 48 h before counting CFU.
DNA extraction
Luminal chyme and mucosal samples were placed in S.T.A.R buffer (Roche) complimented with acid-washed glass beads (size <106 μm, Sigma-Aldrich) directly after dissection. Cells were lysed by homogenization in a MagNaLyser (Roche) at 6500 rpm for 2 × 20 s with a cooling step in-between. Samples were centrifuged at 14,000 g for 5 min. The supernatants were transferred to 96-well plates and DNA was extracted using the Mag Mini LGC kit (LGC Genomics, Teddington, UK) according to the manufacturer’s protocol in a KingFisher Flex DNA extraction robot (Thermo Fisher Scientific).
Real-time quantitative
Mucosal samples were placed in RNAlater (Sigma-Aldrich) directly after dissection. mRNA was isolated with NucleoSpin RNA/Protein Purification kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol and cDNA was synthesized with the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). The primers used for mRNA expression were as follows: Gapdh, forward 5′-CTTCAACAGCAACTCCCACTCTT-3′ and reverse 5′-GCCGTATTCATTGTCATACCAGG-3′ (Tm 60 °C); iNOS, forward 5′-GACATTACGACCCCTCCCAC-3′ and reverse 5′-ACTCTGAGGGCTGACACAAG-3′ (Tm 62 °C); NOX1, forward 5′-GTGATTACCAAGGTTGTCATGC-3′ and reverse 5′-AAGCCTCGCTTCCTCATCTG-3′ (Tm 64 °C). The primer used for genomic quantification were as follows: 16S rRNA gene (modified from ref. 30) forward 5′-TAGCTATTACCGCGGCTGCT-3′, and reverse 5′-ACTCCTACGGGAGGCAGCAGT-3′ (Tm 64 °C). Gapdh forward 5′-AATACGGCTACAGCAACAGG-3′ and reverse 5′-TCTCTTGCTCAGTGTCCTTG-3′ (Tm =56 °C). The RT-qPCR was performed with FirePol EvaGreen qPCR Supermix (Solis BioDyne, Tartu, Estonia) in accordance with the manufacturer’s protocol in a LightCycler 480 Instrument II (Roche). The thermal cycle parameters were as follows: 12 min at 95 °C; 40 cycles of 15 s at 95 °C followed by 20 s at Tm (primer optimized); 20 s at 72 °C. LinReg Software was used to calculate Cq values and primer efficiency. The exact efficiency was used for the comparative Cq values analyzes.
16S rRNA gene sequencing
The analysis of the composition of the gut microbiota was performed on iNOS and NOX1 KO and their respective WT mice. The 16S rRNA gene sequence workflow analysis has previously been reported.31 Briefly, after DNA extraction the 16S rRNA gene was PCR amplified for 25 cycles using prokaryotes-targeting primers developed by Yu et al.32 The PCR product was purified with AMPure XP (Beckman-Coulter, Brea, CA) and 10 further PCR cycles were performed. The resulting amplicons were sequenced on Illumina MiSeq V3 platform (Illumina, San Diego, CA). Resulting 300 bp paired-end reads were further paired-end joined, quality-filtered using QIIME33 and clustered with 97% identity level using closed-reference usearch v7.0 algorithm34, 35 against Greengenes database v13.8.36
Statistical analyses
Statistical significance values were calculated in the GraphPad Prism software (La Jolla, CA). Averages are presented as mean and variances as standard error of the mean (s.e.m.). Statistical significance level was set as α=0.05. We chose 3,000 sequences per sample as a cut-off to normalize the sequencing data. Linear discriminant analysis effect size (LefSe) with LDA score >2 was used to identify taxa associated with the KO groups.37 PCA were performed in Unscrambler 14.1 (Camo software, Oslo, Norway).
References
Larsson, E. et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61, 1124–1131 (2012).
Ha, E.M., Oh, C.T., Bae, Y.S. & Lee, W.J. A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 (2005).
Chavez, V., Mohri-Shiomi, A. & Garsin, D.A. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect. Immun. 77, 4983–4989 (2009).
Moskwa, P. et al. A novel host defense system of airways is defective in cystic fibrosis. Am. J. Respir. Crit. Care Med. 175, 174–183 (2007).
Grasberger, H., El-Zaatari, M., Dang, D.T. & Merchant, J.L. Dual oxidases control release of hydrogen peroxide by the gastric epithelium to prevent Helicobacter felis infection and inflammation in mice. Gastroenterology 145, 1045–1054 (2013).
Grasberger, H. et al. Increased Expression of DUOX2 Is an Epithelial Response to Mucosal Dysbiosis Required for Immune Homeostasis in Mouse Intestine. Gastroenterology 149, 1849–1859 (2015).
Jones, R.M. & Neish, A.S. Redox signaling mediated by the gut microbiota. Free Radic. Biol. Med. 105, 41–47 (2017).
Chu, F.F., Esworthy, R.S., Doroshow, J.H. & Shen, B. NADPH oxidase-1 deficiency offers little protection in Salmonella typhimurium-induced typhlitis in mice. World J. Gastroenterol. 22, 10158–10165 (2016).
Pircalabioru, G. et al. Defensive mutualism rescues NADPH oxidase inactivation in gut infection. Cell Host Microbe 19, 651–663 (2016).
Shaked, H. et al. Chronic epithelial NF-kappaB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc. Natl Acad. Sci. USA 109, 14007–14012 (2012).
Lundberg, J.O. & Weitzberg, E. Biology of nitrogen oxides in the gastrointestinal tract. Gut 62, 616–629 (2013).
Daiber, A., Oelze, M., Steven, S., Kroller-Schon, S. & Munzel, T. Taking up the cudgels for the traditional reactive oxygen and nitrogen species detection assays and their use in the cardiovascular system. Redox Biol. 12, 35–49 (2017).
Reikvam, D.H. et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS ONE 6, e17996 (2011).
Laukens, D., Brinkman, B.M., Raes, J., De Vos, M. & Vandenabeele, P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev. 40, 117–132 (2016).
Asghar, M.N. et al. In vivo imaging of reactive oxygen and nitrogen species in murine colitis. Inflamm. Bowel Dis. 20, 1435–1447 (2014).
Han, W., Li, H., Segal, B.H. & Blackwell, T.S. Bioluminescence Imaging of NADPH Oxidase Activity in Different Animal Models. J. Vis. Exp. 68, e3925 (2012).
Kielland, A., Blom, T., Nandakumar, K.S., Holmdahl, R., Blomhoff, R. & Carlsen, H. In vivo imaging of reactive oxygen and nitrogen species in inflammation using the luminescent probe L-012. Free Radic. Biol. Med. 47, 760–766 (2009).
Zangani, M. et al. Tracking early autoimmune disease by bioluminescent imaging of NF-kappaB activation reveals pathology in multiple organ systems. Am. J. Pathol. 174, 1358–1367 (2009).
Radi, R. Peroxynitrite a stealthy biological oxidant. J. Biol. Chem. 288, 26464–26472 (2013).
Zielonka, J., Lambeth, J.D. & Kalyanaraman, B. On the use of L-012, a luminol-based chemiluminescent probe, for detecting superoxide and identifying inhibitors of NADPH oxidase: a reevaluation. Free Radic. Biol. Med. 65, 1310–1314 (2013).
Goiffon, R.J., Martinez, S.C. & Piwnica-Worms, D. A rapid bioluminescence assay for measuring myeloperoxidase activity in human plasma. Nat. Commun. 6, 6271 (2015).
Jones, R.M. et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 32, 3017–3028 (2013).
Sonoyama, K. et al. Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl. Environ. Microbiol. 75, 6451–6456 (2009).
Corcionivoschi, N. et al. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59 (2012).
Leoni, G. et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J. Clin. Investig. 123, 443–454 (2013).
Chooruk, A., Piwat, S. & Teanpaisan, R. Antioxidant activity of various oral Lactobacillus strains. J. Appl. Microbiol. 123, 271–279 (2017).
Miyoshi, A. et al. Oxidative stress in Lactococcus lactis. Genet. Mol. Res. 2, 348–359 (2003).
Deloris Alexander, A., Orcutt, R.P., Henry, J.C., Baker, J. Jr., Bissahoyo, A.C. & Threadgill, D.W. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm. Genome 17, 1093–1104 (2006).
Goodyear, A.W., Kumar, A., Dow, S. & Ryan, E.P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods 405, 97–108 (2014).
Brukner, I., Longtin, Y., Oughton, M., Forgetta, V. & Dascal, A. Assay for estimating total bacterial load: relative qPCR normalisation of bacterial load with associated clinical implications. Diagn. Microbiol. Infect. Dis. 83, 1–6 (2015).
Avershina, E. et al. Transition from infant- to adult-like gut microbiota. Environ. Microbiol. 18, 2226–2236 (2016).
Yu, Y., Lee, C., Kim, J. & Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Edgar, R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
DeSantis, T.Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
We would like to thank Inga Leena Angell for technical support. This work was supported by grants from the EU consortium DiMI (LSHB-CT-2005-512146), the Norwegian University of Life Sciences, the Norwegian Cancer Society, and the Norwegian Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
A.K. conceived and designed the study. C.M. and A.K. performed most of the experiments and analyzed data. S.D.C.R. and O.A.H. performed experiments. K.R. provided equipment and reagents. C.M., H.C., and A.K. wrote the paper. All authors reviewed and provided comments to the paper.
Rights and permissions
About this article
Cite this article
Matziouridou, C., Rocha, S., Haabeth, O. et al. iNOS- and NOX1-dependent ROS production maintains bacterial homeostasis in the ileum of mice. Mucosal Immunol 11, 774–784 (2018). https://doi.org/10.1038/mi.2017.106
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2017.106
This article is cited by
-
Metabolic network of the gut microbiota in inflammatory bowel disease
Inflammation and Regeneration (2024)
-
Transcriptional analysis reveals specific niche factors and response to environmental stresses of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents
BMC Microbiology (2021)
-
Colitis susceptibility in mice with reactive oxygen species deficiency is mediated by mucus barrier and immune defense defects
Mucosal Immunology (2019)
-
NADPH oxidases and ROS signaling in the gastrointestinal tract
Mucosal Immunology (2018)