Abstract
T helper type (Th17) cytokines such as interleukin (IL)-17A and IL-22 are important in maintaining mucosal barrier function and may be important in the pathogenesis of inflammatory bowel diseases (IBDs). Here, we analyzed cells from the colon of IBD patients and show that Crohn’s disease (CD) patients had significantly elevated numbers of IL-17+, CD4+ cells compared with healthy controls and ulcerative colitis (UC) patients, but these numbers did not vary based on the inflammatory status of the mucosa. By contrast, UC patients had significantly reduced numbers of IL-22+ cells in actively inflamed tissues compared with both normal tissue and healthy controls. There was a selective increase in mono-IL-17-producing cells from the mucosa of UC patients with active inflammation together with increased expression of transforming growth factor (TGF)-β and c-Maf. Increasing concentrations of TGF-β in lamina propria mononuclear cell cultures significantly depleted Th22 cells, whereas anti-TGF-β antibodies increased IL-22 production. When mucosal microbiota was examined, depletion of Th22 cells in actively inflamed tissue was associated with reduced populations of Clostridiales and increased populations of Proteobacteria. These results suggest that increased TGF-β during active inflammation in UC may lead to the loss of Th22 cells in the human intestinal mucosa.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) is characterized by chronic inflammation in the human intestinal tract and is comprised of two diseases: Crohn’s disease (CD) and ulcerative colitis (UC).1 Although both CD and UC are thought to be driven by an aberrant immune response against intestinal flora and/or dietary antigens, their pathophysiology is quite different.2, 3 Classically, CD was characterized as a predominantly T helper type 1 (Th1)-mediated inflammatory state and UC a Th2-mediated inflammatory state.1 However, recent evidence has increasingly shown a role for Th17 cells in the pathogenesis of the disease.4 Th17 cells are a subset of CD4+ helper T cells that produce interleukin (IL)-17A, IL-17F, and IL-22 and are enriched at mucosal sites.5 The cytokines produced by Th17 cells are elevated in the serum and intestinal tissue of patients with IBD, and thus Th17 cells may have a role in IBD pathogenesis.4 Elevated numbers of activated Th17 cells have been found in the colon and blood of CD patients.6 However, in humans not all Th17 cells produce IL-22, and a Th22 subset of CD4+ helper T cells that produces IL-22, but not IL-17 has been identified.7 Although IL-17 promotes recruitment and activation of neutrophils, IL-22 promotes mucosal healing through epithelial proliferation and increased mucus production.8, 9 Although IL-17- and IL-22-producing CD4+ helper T cells are clearly important in regulating mucosal barrier function and intestinal homeostasis, their exact role in the pathogenesis of CD and UC is still unclear.4 These cytokines can also be produced by other lymphocytes, such as NK cells and innate lymphoid cells.10, 11
In many IBD studies, the gut specimens used for isolating lamina propria mononuclear cells (LPMCs) come from patients undergoing bowel resection for IBD, which reflects a condition of severe disease that has failed treatment. A more dynamic perspective of varying inflammatory states in IBD could be obtained through the analysis of pinch biopsy material taken during endoscopy. Recently, we characterized LPMCs from pinch biopsies taken from multiple locations of the colon of an UC patient that had self-infected himself with Trichuris trichiura to treat his own symptoms.12 During active colitis, inflamed mucosa was infiltrated with CD4+ T cells producing only IL-17, whereas mucosal healing and worm-colonized tissues were associated with CD4+ T cells producing IL-22. Now, in this study, we analyzed LPMCs from pinch biopsies taken from CD and UC patients, using pathological data from paired mucosal biopsies to inform us of the inflammatory state of the biopsied site.
We find that CD4+ T cells that are producing only IL-17 are enriched in inflamed portions of the colon of UC patients, and this is associated with a relative decline in Th22 cells producing only IL-22. Transforming growth factor (TGF)-β is a pleiotropic cytokine with complex biology.13 In addition to inducing FoxP3(+) (forkhead box p3) T regulatory (Treg) cells,5 it is involved in many other cellular processes.13 In the presence of inflammatory cytokines such as IL-6, IL-23, and IL-1b, TGF-β has been shown to promote Th17 differentiation.5 However, it was shown recently that TGF-β could inhibit IL-22 production in mice through the transcription factor c-Maf.14 We find here that TGF-β can also inhibit differentiation of IL-22-producing human CD4+ cells in LPMCs.
Alterations in the mucosal microbiota have been previously observed in UC patients.15, 16 By comparing immunological data generated by flow cytometry and microbial composition data generated by 16S ribosomal deep sequencing, we find that the reduced abundance of Th22 and also Th1 cells in actively inflamed tissue is associated with specific alterations to the bacterial communities of the mucosal microbiota.
Results
Increased Th17 and decreased Th22 cells are found in LPMCs from the inflamed mucosa of UC patients
In this study, we examined the relative abundance of IL-17- and IL-22-producing CD4+ T cells in the intestinal mucosa of IBD patients. We isolated LPMCs from pinch biopsies taken from areas of inflamed and non-inflamed tissue from 12 UC and 13 CD patients, as well as from the normal colon of 27 healthy individuals, who served as non-inflammatory controls. By gating on live CD4+ T cells within the CD3+ population (Figure 1a), we observed elevated proportions of Th17 cells (both IL-17+, IL-22− and IL-17+, IL-22+) in LPMCs of CD patients compared with both UC patients and healthy controls (Figure 1b). By contrast, there was a significant decrease in the percentage of Th22 cells (IL-22+, IL-17−) in the LPMCs of UC patients, but not in CD patients, when compared with healthy controls. This suggests that Th17 cells are increased in the intestinal mucosa of CD patients, whereas Th22 cells are decreased in UC patients relative to healthy individuals.
T helper type 17 (Th17) cells are enriched in Crohn’s disease (CD) patients and Th22 cells are depleted in ulcerative colitis (UC) patients. (a) Representative gating strategy for the FACS (fluorescence-activated cell sorter) analysis of lamina propria mononuclear cells (LPMCs) isolated from a CD patient, showing live CD3+ CD4+ lymphocytes with intracellular staining for interleukin (IL)-17 and IL-22 after phorbol 12-myristate 13-acetate/Ionomycin stimulation. (b) Representative FACS plots showing differences in intracellular IL-17 and IL-22, gated on CD4+ cells (as shown in a), between healthy subjects, UC patients, and CD patients. (c) Scatterplots summarizing the percentages of CD4+ cells that are IL-17+ and IL-22+ (as shown in b) from LPMCs of healthy subjects, UC patients, and CD patients. FSC, forward scatter; SSC, side scatter. *P<0.05; **P<0.005.
As the colon of IBD patients vary considerably in terms of their inflammatory states, we compared our flow cytometry results with the pathology results obtained from paired pinch biopsies to further examine the role of Th17 and Th22 cells during different states of inflammation. The pathology results subdivided our pinch biopsies into three degrees of inflammation: normal/no inflammation, inactive chronic colitis, and active chronic colitis. Active chronic colitis is defined by neutrophil infiltration into the epithelium in the setting of epithelial cell damage, whereas inactive chronic colitis refers to the increase of mononuclear cells in the lamina propria in the presence of structural changes to the mucosa.17 We observed no significant differences in the percentage of CD4+ IL-17+ or IL-22+ cells in CD patients based on the inflammatory status of the mucosa (Figure 2a). However, in UC patients, there was a significant increase in the percentage of CD4+ IL-17+, IL-22− cells in tissue with active chronic colitis compared with both inactive chronic colitis and normal tissue (Figure 2c). This increase was further accompanied by a relative decline in the percentage of CD4+ IL-22+, IL-17− cells in the actively inflamed mucosa.
T helper type 17 (Th17) cells are enriched and Th22 cells are reduced in the regions of the colon with active inflammation in patients with ulcerative colitis (UC) but not Crohn’s disease (CD). (a) Representative FACS (fluorescence-activated cell sorter) plots showing intracellular interleukin (IL)-17 and IL-22, gated on CD4+ cells (as shown in Figure 1), from lamina propria mononuclear cells (LPMCs) of CD patients isolated from regions with normal, inactive, or active colitis. (b) Scatterplots summarizing the percentages of CD4+ cells that are IL-17+ and IL-22+ (as shown in a) from LPMCs of CD patients. There are no statistically significant differences between any of the groups. (c) Representative FACS plots showing differences in intracellular IL-17 and IL-22 between regions of the large intestine that are considered normal, with inactive colitis, or active colitis based on histopathology from UC patients. (d) Scatterplots summarizing the percentages of CD4+ cells that are IL-17+ and IL-22+ (as shown in c) from LPMCs of UC patients isolated from regions with normal, inactive, or active colitis. *P<0.05; **P<0.005.
Active inflammation in the mucosa of UC patients alters the composition of cytokine-producing intestinal lymphocytes
Our flow cytometry panels included IFNγ, TNFα, and IL-4, in addition to IL-17 and IL-22. To determine whether changes in the percentages of Th1 and Th2 cytokines were also occurring with increased inflammation, we looked at the relative compositions of IFNγ, TNFα, and IL-4 production in the intestinal mucosa of our IBD cohort through boolean gate analysis (Figure 3a). Analysis of the 32 boolean gates produced from our five cytokines of interest revealed a decrease in cytokine combinations involving IFNγ+ cells and an increase in TNFα+ combinations with increased mucosal inflammation in our UC cohort (Figure 3b). As previously discussed, actively inflamed tissue in UC patients also exhibited an increase in combinations involving IL-17+ cells and a decrease in combinations involving IL-22 cells. No significant changes in the combinations involving IL-4+ cells were observed. In CD patients, the main difference in the boolean gate combinations of our five cytokines involved an increase in IL-17+ combinations in tissue with active colitis compared with normal tissue (Figure 3c).
Poly- and mono-cytokine producing CD4+ cells analyzed by boolean gate analysis from the regions of the large intestine of ulcerative colitis (UC) and Crohn’s disease (CD) patients that are normal, with inactive colitis, or active colitis. (a) Representative boolean gating strategy for interleukin (IL)-17, IL-22, interferon-γ (IFNγ), tumor necrosis factor-α (TNFα), and IL-4 in CD4+ lamina propria mononuclear cells (LPMCs) of a UC patient. (b) Pie charts showing the averaged pattern of cytokine production in CD4+ cells from the regions with normal, inactive, and active inflammation of UC patients. Each slice within the pie chart represents a specific combination of cytokine staining. (c) Pie charts showing the averaged pattern of cytokine production in CD4+ cells from the regions with normal and active inflammation of CD patients. Each slice within the pie chart represents a specific combination of cytokine staining. (d) Scatter plots summarizing the percentages of CD4+ cells that are only producing one cytokine (Mono IL-17, IL-22, IFNγ, TNFα, and IL-4) from LPMCs of healthy subjects and UC patients isolated from the regions with normal, inactive, or active colitis. *P<0.05; **P<0.005. NS, not significant; SSC, side scatter.
As we had previously found that active colitis was associated with an increased percentage of mono-IL-17+ cells,12 we looked at other mono-cytokine-producing CD4+ cells in LPMCs of our UC cohort. Consistent with our previous studies, we observed an increase in mono-cytokine-producing IL-17+ CD4+ T cells in tissue with active chronic colitis compared with both the normal tissue of UC patients and of healthy controls. This increase in mono-IL-17+ cells was accompanied by an increase in CD4+ cells producing only TNFα in actively inflamed tissue. By contrast, we observed a selective decrease in the percentage of both mono-IL-22+ and mono-IFNγ+ cells in the actively inflamed mucosa compared with the normal tissue of UC patients and of healthy controls. No significant differences were observed in the percentage of mono-IL-4+ cells in UC patients based on the inflammatory status of the mucosa (Figure 3d).
We expanded our boolean gates to include all possible 2, 3, 4, and 5 cytokine combinations of our five cytokines of interest, resulting in 232 boolean combinations. Through unsupervised hierarchical clustering, we found that these boolean gates were able to separate out the normal and active chronic colitic tissue of our UC patients into two distinct clusters, with the inactive chronic colitic tissue interspersed between these two groups (Figure 4). In CD patients, however, the 232 boolean gates were not able to differentiate the patients based on the inflammatory status of the mucosa (Figure 4). Instead, the clustering of the boolean gate analysis seemed to be driven by individual differences found between patients. These results suggest that the inflammatory response in mucosa of UC and CD are quite different, whereby the inflammatory state of tissues from UC patients determines the type of inflammatory CD4+ cells there, whereas in CD patients the systemic differences in immune responses between patients is more important in determining the type of inflammatory CD4+ cells in the mucosa than the inflammatory state of the mucosal tissues. Increased proportions of IFNγ+ and IL-22+ cells were found in normal tissue compared with actively inflamed tissue in UC patients, and increased TNFα in combination with these two cytokines was also observed (Supplementary Figure S1). In the actively inflamed tissue of UC patients, there was an increase in IL-17+ and TNFα+ cells compared with normal tissue (Supplementary Figure S2). These observations are consistent with previous studies showing that IL-22 expression by human CD4+ T cells correlates better with expression of the Th1 cell subset markers IFNγ and T-bet than with expression of the Th17 cell subset marker IL-17A.18, 19
Unsupervised hierarchical clustering analyses of the combinations of polycytokine-producing CD4+ cells in ulcerative colitis (UC) patients and Crohn’s disease (CD) patients. Lamina propria mononuclear cells (LPMCs) from regions with active inflammation in UC patients have similar compositions of polycytokine-producing CD4+ cells and are tightly clustered, but for CD patients, LPMCs from regions with active inflammation are more disparate. Boolean gate combinations for every possible 2–5 cytokine combinations of interferon-γ, tumor necrosis factor-α, interleukin (IL)-4, IL-17, and IL-22 were extracted from FACS (fluorescence-activated cell sorter) analysis of intracellular cytokine-stained CD4+ cells from the LPMCs of (a) UC and (b) CD patients isolated from regions with normal, inactive, or active colitis. The extracted data were then subjected to unsupervised clustering analysis to identify samples isolated from normal (green), inactive (purple) or active (red) colitis regions, that had related effector CD4+ populations and to determine the relationship between effector CD4+ populations. Blue=less than the mean; yellow=greater than the mean; black=mean values. This analysis identified two clusters of effector CD4+ populations that separated samples from tissues with active colitis and normal tissue from UC patients. Cluster 1 represents a group of boolean gate combinations that are more abundant in the normal and inactive chronic colitis tissues of UC patients compared with actively inflamed mucosa, and cluster 2 represents a cluster of boolean gate combinations more abundant in active colitis tissues of UC patients compared with normal and inactive chronic colitis tissues.
TGF-β can reduce the number of IL-22-producing lamina propria CD4+ T cells in vitro
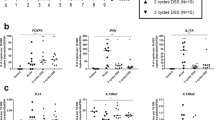
Recently, increasing concentrations of TGF-β was shown to suppress IL-22 production in mouse T cells through the transcription factor c-Maf.14 We performed real-time PCR analysis on paired pinch biopies taken from the UC patients that we had analyzed by flow cytometry for expression of TGF-β, c-Maf, and IL-17. This was compared with biopsies from healthy controls (Figure 5). As expected, IL-17 was significantly upregulated in tissues with active inflammation compared with normal and inactive inflammation. TGF-β transcript expression was also significantly higher in actively inflamed tissues compared with normal tissue and healthy controls. Expression of c-Maf was higher in the UC biopsies compared with healthy controls but was not significantly different between biopsies with active inflammation and normal tissue. Additionally, we found an increased expression of IL13RA2, MMP7 (matrix metalloproteinase 7), CHI3L1 (chitinase-3-like protein 1), and FoxP3 in actively inflamed tissue compared with tissue from healthy controls. Although no significant differences were seen between normal tissue and actively inflamed tissue for these markers, their expression did trend higher in actively inflamed tissue (Figure 5). We also examined cytokines involved in Th17 and Th22 differentiation and found that IL-6 and IL-23 were increased in actively inflamed tissue vs. normal tissue, and IL-1b trended towards a higher expression in active tissue (Supplementary Figure S3). IL-22 expression levels by real-time PCR were below the limit of detection among most samples (data not shown). We therefore examined expression of genes known to be induced by IL-22,20 such as the antimicrobial S100A peptides and Reg3 proteins (Supplementary Figure S4). Expression of these genes was mostly not significantly different between normal tissue and actively inflamed tissue (Supplementary Figure S4). However, many of these IL-22 target genes were identified in mice and regulation in humans is unclear. Additionally, these genes could be induced by IL-17 making it difficult to distinguish between the effects of IL-17 and IL-22. By querying a gene expression–profiling experiment performed on human bronchial epithelial cells to study the response to IL-22 and IL-17,21 we identified CCL5 (C-C motif chemokine ligand 5) as a gene that may be induced by IL-22 more significantly than IL-17 (Supplementary Figure S4). Real-time PCR analysis of CCL5 showed a reduced expression of CCL5 in tissue from UC patients compared with healthy controls (Figure 5), although this was not significantly different between actively inflamed tissue and inactive or normal tissue from UC patients. Additional studies are needed to determine whether this is an indication of functional IL-22 deficiency.
Expression levels of genes important in mucosal immunity and T-cell differentiation. Expression levels of interleukin (IL)-17A, TGF-β1 (transforming growth factor β1), c-Maf, IL13RA2, MMP7 (matrix metalloproteinase 7), CHI3L1 (chitinase-3-like protein 1), FoxP3 (forkhead box p3), and CCL5 (C-C motif chemokine ligand 5) were measured by reverse transcriptase–PCR analysis of pinch biopsies taken from healthy individuals and in normal, inactive, and active tissue of UC patients. All cycle threshold values were normalized to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) values and log transformed. *P<0.05; **P<0.005; ***P<0.0005.
To determine whether TGF-β can suppress IL-22 production in human CD4+ cells under activating conditions, we cultured peripheral blood mononuclear cells (PBMCs) and LPMCs from healthy individuals with increasing concentrations of TGF-β for 6 days in the presence of IL-6, IL-1b, IL-23, and anti-CD3/CD28 beads (Figure 6). We then analyzed the generation of IL-17- and IL-22-producing CD4+ cells by intracellular cytokine staining (Figure 6a) and the differentiation of FoxP3+ cells by intranuclear staining (not shown). In LPMCs, we observed that increasing concentrations of TGF-β suppressed CD4+ IL-22+, IL-17− cells in culture, while increasing CD4+ CD25+ FoxP3+ Treg cells (Figure 6a). Blocking antibodies against TGF-β did the opposite and increased the frequency of CD4+ IL-22+, IL-17− cells and reduced the frequency of FoxP3+ Tregs. We observed no significant differences in CD4+ IL-17+ production with increasing concentrations of TGF-β. Similar results were observed in PBMC cultures from healthy individuals (Figure 6b). These results confirm in human LPMCs, previous findings in mice that TGF-β can suppress IL-22 production.
Transforming growth factor (TGF)-β inhibits the differentiation of CD4+ interleukin (IL)-22+ cells from lamina propria mononuclear cells (LPMCs) and peripheral blood mononuclear cells (PBMCs) of healthy individuals. (a, b) Representative FACS (fluorescence-activated cell sorter) plot of intracellular IL-22 and IL-17 staining of CD4+ cells from LPMCs (a) or PBMCs (b) after 6 days of culture in increasing concentrations of TGF-β or with a blocking antibody against TGF-β in activation conditions that include anti-CD3/anti-CD28 beads, IL-2 (10 ng ml−1), IL-6 (10 ng ml−1), IL-23 (10 ng ml−1), and IL-1b (10 ng ml−1). (c, d) Bar charts summarizing the changes to the proportions of T helper type 17 (Th17), Th22, and regulatory T cells after 6 days of LPMC (c) or PBMC (d) cultures under activating conditions (as described for a and b) from LPMCs of n=6 individuals and PBMCs from n=4 individuals. Error bars show the variation between individuals. Changes are normalized against the percentage of cells producing cytokines at 0 ng ml−1 TGF-β as being 100%.
Alterations to the mucosal microbiota are associated with active inflammation
Changes to bacterial taxa may be linked with disease pathogenesis in IBD patients.2, 4, 15, 22 To investigate the mucosal microbiota communities in biopsies from UC patients using culture-independent methods, we performed deep sequencing analysis on the variable region 4 (V4) region of bacterial 16S rRNA.23 We found that the phylum Firmicutes was more abundant in normal tissue compared with actively inflamed tissue, whereas the phylum Proteobacteria was more abundant in the inflamed mucosa (Figure 7a). We confirmed these results using the LEfSe (linear discriminant analysis (LDA) effect size) method,24 defining normal, inactive, and active inflammation as classes of microbial communities. The increase in Firmicutes in normal tissue was mostly due to differences in the genus Faecalibacterium, whereas the increase in Proteobacteria in active inflammation was mostly due to differences in the genus Acinetobacter (Figure 7b). The abundance of these clades was also significantly different between normal and actively inflamed tissue for the phylum Firmicutes down to the family Ruminococcaceae and for the phylum Proteobacteria down to the genus Acinetobacter (Figure 7c).
Correlations of the mucosal microbiota communities with inflammation status in patients with ulcerative colitis. (a) Relative abundance of the taxonomic groups for biopsy samples from each tissue type. The eight most abundant phyla over all samples are presented. (b) Histogram of linear discriminant analysis (LDA) scores computed for features differentially abundant between normal and actively inflamed tissues. Features with LDA scores >0.02 are presented, and taxonomic level is indicated by p (phylum), c (class), o (order), f (family), and g (genus). (c) Relative abundance of specific phyla and genera in a and b that were differentially abundant in different tissue types. Each point represents a biopsy sample, and mean is indicated with a horizontal line. The effect of tissue on abundance was determined using a Kruskal–Wallis test (Firmicutes, P=0.045; Faecalibacterium, P=0.042; Proteobacteria, P=0.014; Acinetobacter, P=0.015). Differences between individual tissues were determined using Dunn’s Multiple Comparison Test, and significant differences (P<0.05) are indicated with an asterisk. (d) A Spearman’s correlation was performed between the relative abundance of Firmicutes, Clostridiales, Proteobacteria, and Acinetobacter and the percentage of CD4+ interleukin (IL)-22+, IL-17− cells. These correlations were all significant (P<0.05) with P values and r values shown on graphs. Green=normal; blue=inactive; red=active.
We also analyzed microbial diversity within samples (α-diversity) and between samples (β-diversity). Shannon’s diversity index was slightly higher for normal tissue than for inflamed tissue, although not statistically significant (Supplementary Figure S5A,B). We observed slight clustering of normal and actively inflamed tissue by Principal Coordinate Analysis and Unweighted Pair Group Method with Arithmetic Mean trees (Supplementary Figure S5C,D). Normal and active inflammation separated slightly along the PC1 axis, which corresponded with the separation of Firmicutes and Proteobacteria.
We were next interested to see whether there was a correlation between the frequency of Th22 cells and the relative abundance of Firmicutes, Ruminococcaceae, Proteobacteria, and Acinetobacter in our UC cohort. We found that the percentage of CD4+ IL-22+, IL-17− cells was positively associated with the phylum Firmicutes (P=0.0143), down to the order Clostridiales (P=0.0397) and negatively associated with the phylum Proteobacteria (P=0.0311) down to the genus Acinetobacter (P=0.0492) (Figure 7d). When we examined the relationship with IFNγ+ Th1 cells, we also observed a positive association with the order Clostridiales that was almost significant (P=0.0545) and a negative association with the genus Acinetobacter (P=0.0430) (Supplementary Figure S6A). This is consistent with IL-22 expression correlating with expression of IFNγ.18, 19 We found no significant correlations between the frequency of Th22 cells and the relative abundance of the family Ruminococcaceae and genus Faecalibacterium within the Firmicutes phylum (Supplementary Figure S6B). We also examined the correlation between the percentage of CD4+ IL-17+, IL-22− cells and these bacteria taxa and found no significant correlations between these parameters (Supplementary Figure S6C).
Discussion
Active inflammation during UC is associated with disruption of the mucus barrier and an aberrant response against the gut microbiota.2, 15 In this study, we showed that actively inflamed tissue from UC patients (but not CD patients) was specifically associated with an increased percentage of mono-IL-17-producing Th17 cells and a depletion of Th22 cells compared with normal and healthy tissue. The increased ratio of Th17/Th22 cells during active inflammation in UC may explain the manifestation of active inflammation as neutrophilic superficial lesions, whereas CD is characterized by deeper ulcers, granulomas and mononuclear cell–dominated lesions and associated with a more systemic increase in Th17 cells that does not vary based on inflammatory status. The depletion of Th22 cells we see during active inflammation in UC is strikingly similar to a recent report that Th22 cells were depleted during chronic HIV infection, when there was increased microbial translocation and damage to epithelial integrity.25 However, it is important to note that the loss of Th22 cells during active inflammation is simply a correlation and whether this depletion is important for disease pathogenesis is not known. Additional studies are required to demonstrate causality in UC pathogenesis. Still, these results are consistent with a protective role for IL-22 in the gut. As IL-22, a member of the IL-10 cytokine family, can enhance epithelial regeneration and regulate tissue repair, its protective role for the intestinal mucosa has also been demonstrated using several mouse models of colitis.26, 27, 28 Although previous reports did not identify significant changes in Th17 or Th22 cells in UC,29 these differences may be observed here because we are analyzing pinch biopsies with paired pathology samples that inform us of the inflammatory status of the biopsies that we have collected.
An important caveat to this study is that the Th22 cells are detected by intracellular cytokine staining after phorbol 12-myristate 13-acetate (PMA)/Ionomycin stimulation, and it is not certain that these cells are actually producing IL-22 within the intestinal tissues in vivo. It is therefore unclear whether production of IL-22 is actually reduced during active inflammation in UC. Indeed, a previous study found increased IL-22 by reverse transcriptase–PCR analysis of UC tissues.30 It was, however, previously demonstrated that IL-22 secretion, as detected by enzyme-linked immunosorbent assay of supernatants from purified T cells, is consistent with IL-22 detection by intracellular cytokine staining of CD4+ cells.18, 31 It will be important in the future to characterize Th22 cells in the human mucosa in response to stimulation with specific antigens (e.g., to commensal bacteria).
Despite these limitations, our studies support an emerging consensus that Th22 cells are an important component of protecting the mucosal barrier in human disease as well as in mouse models of colitis. Recently, an important study using various mutant mice dissected distinct roles for Th22 cells and IL-22-producing innate lymphocytes (innate lymphoid cells) during Citrobacter rodentium infection.32 Although IL-22+ innate lymphoid cells are important early during infection, IL-23-independent Th22 cells induced by IL-6 have a critical role during the adaptive phase of the host response against C. rodentium.32 These authors also confirmed the previous report14 that TGF-β significantly suppressed Th22 cell differentiation. Although Th22 cells have been described mainly in humans, it is difficult to demonstrate an in vivo functional role for these cells through clinical studies. By performing adoptive transfer experiments, the study by Basu et.al.32 can clearly demonstrate an important functional role for Th22 cells during adaptive phases of a response in the intestinal mucosa. Our study, as well as a study showing depletion of Th22 cells in HIV patients,25 provides evidence that the mouse studies may be physiologically relevant to human intestinal diseases.
We identify a possible mechanism to explain the preferential loss of Th22 cells in actively inflamed tissue, based on studies showing that TGF-β suppresses IL-22 production while inducing IL-17A in T cells under activating conditions via c-Maf.14 Although transcriptional expression of TGF-β may not accurately reflect levels of active TGF-β in tissue,33 we did find that, at a transcriptional level, TGF-β was more highly expressed in inflamed tissue. In the future, to accurately determine whether there is more active TGF-β in actively inflamed UC tissue, biopsies will have to be cultured ex vivo for bioassay analysis of supernatants. It is possible that increased TGF-β activity is a reflection of inflammation in general that is not specific to UC. Hence, we currently lack a direct cause and effect relationship between expression of TGF-β transcripts and the observed loss of Th22 cells.
We were able to replicate the previous mouse studies14 using LPMCs isolated from the human colon to show that increasing concentrations of TGF-β suppressed IL-22 production under Th17 activation conditions, whereas blocking antibodies against TGF-β increased IL-22 production. However, as we did not isolate naive T cells from LPMCs, we may only be assessing the production of IL-22 from pre-existing Th22 cells. TGF-β is generally considered to have a protective role against intestinal inflammation by inducing FoxP3(+) Treg cells,34 but it is also a pleiotropic cytokine with complex biology.13 Although high levels of active TGF-β are produced in the inflamed tissues of IBD patients,34 it is not sufficient to stop mucosal inflammation. Perhaps TGF-β suppresses both a protective IL-22 response as well as pathogenic inflammatory responses. Perhaps it may be possible in the future to develop a more specific inhibitor (e.g., against c-Maf) that would be able to restore Th22 cells in the lamina propria and promote mucosal healing, without impacting Tregs. Future studies are needed to directly document the induction of c-Maf and investigate its role in human LPMCs, as well as naive CD4+ T cells, in mediating the effects of TGF-β on suppressing the differentiation of Th22 cells.
Alterations in the gut microbiota has been noted previously in IBD patients,35, 36 although most of these studies do not investigate specific differences in the inflammatory status of the biopsied tissue. We find in this study that there are specific alterations in the mucosal microbiota of UC patients that are associated with the inflammatory status of the tissue. Consistent with previous reports, we see increased relative abundance of the phylum Proteobacteria in tissues with active inflammation and decreased relative abundance of the phylum Firmicutes, in particular of the order Clostridiales. Additionally, we were able to directly compare the relative frequencies of Th17 and Th22 cells with the composition of the mucosal microbiota. We found a positive correlation between the percentage of Th22 and the phylum Firmicutes down to the order Clostridiales and a negative correlation with the phylum Proteobacteria down to the genus Acinetobacter. These associations were also observed for IFNγ+ Th1 cells; hence, these correlations with the mucosal microbiota are not specific only to the frequency of Th22 cells.
In addition to Th22 cells, mucosal innate lymphocytes are another major source of IL-22.10, 11, 29 The flow cytometry panels used in this study are not appropriate for characterizing innate lymphocytes. However, we have tried modifying our gating strategy to select for all of the IL-22+ cells in the live lymphocyte cell gate and determine whether there are significant non-CD4+ sources of IL-22 such as innate lymphocytes and we find too few CD3− IL-22+ cells for proper quantification. Surgical specimens may be required for characterizing IL-22-producing innate lymphocytes.
In conclusion, our results are consistent with the hypothesis that IL-22 and Th22 cells in the intestinal mucosa may have a key role in maintaining mucosal barrier function. The depletion of Th22 cells during active inflammation in UC patients may be associated with the disruption of mucosal epithelial integrity. TGF-β may have a role in this depletion by suppressing IL-22 production in T cells from the intestinal mucosa, and this depletion is associated with specific alterations to the mucosal microbiota, namely with increased abundance of Proteobacteria and decreased abundance of Firmicutes. Identifying strategies to restore Th22 cells to the intestinal mucosa during active inflammation may be an avenue for novel therapeutics against UC.
Methods
Isolation of cells. Pinch biopsies were taken from IBD patients and healthy subjects undergoing surveillance colonoscopies at the Mount Sinai Medical Center, New York, NY, USA and the Mahattan VA Hospital, New York, NY, USA. Institutional review board approval was obtained before involving patients in the study. Pinch biopsies from the colonic mucosa of healthy individuals and IBD patients were digested at 37 °C for 1 h in 100 units ml−1 collagenase Type VIII (Sigma, St Louis, MO) and 0.150 mg ml−1 DNase (Sigma) in complete RPMI 1640 (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum, 2 mM L-glutamine, 100 units ml−1 penicillin, 0.1 mg ml−1 streptomycin, and 0.05 mM 2-Mercaptoethanol. Cells were filtered through a 50-micron filter, washed with 5 ml phosphate-buffered saline (PBS), and pelleted. Cells were then resuspended in 40% Percoll (GE Healthcare, Fairfield, CT) in complete RPMI, under-layed with 80% Percoll, and centrifuged at 2,200 r.p.m. for 20 min at room temperature. LPMCs were collected at the interface and used for subsequent flow cytometry analyses.
FACS (fluorescence-activated cell sorter) staining. Cells were stimulated with 50 ng ml−1 PMA and 500 ng ml−1 Ionomycin for 4 h at 37 °C in the presence of brefeldin A (GolgiPlug, BD, Franklin Lakes, NJ). Following this in vitro stimulation, cells were stained with anti-CD3, anti-CD4, anti-CD8, and anti-CD56 and fixed in 4% paraformaldehyde in PBS. Cells were permeabilized in Perm/Wash buffer (BD) and stained with anti-IFNγ, anti-TNFα, anti-IL-17A, anti-IL-22, and anti-IL-4. Alternatively, staining for FoxP3 was performed using the Fix/Perm buffer kit (eBioscience, San Diego, CA) for fixation and permeabilization. Cells were acquired on an LSRII (BD) and analyzed with FlowJo (Tree Star, Ashland, OR) software. Positive gates for cytokine production are drawn based on control experiments performing Fluorescence Minus One (FMO) controls with PBMC samples stained with the same intracellular cytokine panels.
16S rRNA analyses. DNA was isolated from pinch biopsy material. The V4 region of the 16S rRNA gene was PCR amplified for sequencing with region-specific barcoded primers37 and sequenced on a MiSeq sequencer23 along with other barcoded samples. Reads shorter than 140 bp were discarded. The QIIME suite of analysis tools was used to filter and analyze the sequence data.38 Sequences were assigned to operational taxonomic units (OTUs) with a threshold of 97% pair-wise identity and classified taxonomically according to the Ribosomal Database Project (RDP). Relative abundance of microbial phyla was calculated using the proportion of OTUs present in each tissue sample. A representative sequence for each OTU was aligned using PyNAST and used to build a phylogenetic tree for α-diversity and β-diversity measurements. α-Diversity was calculated using the Shannon index. β-Diversity was calculated using unweighted UniFrac distances between samples and visualized using Principal Coordinate Analysis plots and Unweighted Pair Group Method with Arithmetic Mean trees. We then used the LEfSe algorithm24 through the Galaxy Framework39, 40 to identify significant differences in bacterial taxa between groups. LEfSe uses the Kruskal–Wallis (KW) rank sum test to detect features with significantly different abundances between assigned classes and performs LDA to estimate the effect size of each feature. We chose an LDA score threshold of 0.02.
In vitro culture conditions. Mononuclear cells were harvested either from whole blood using Histopaque solution (Sigma-Aldrich, St Louis, MO) or from colonic pinch biopsies using a Percoll (GE Healthcare) gradient. Mononuclear cells were washed with 1 × PBS and resuspended in sterile RPMI containing 10% fetal bovine serum (Gemini, West Sacramento, CA), 1 μM sodium pyruvate (Gibco, Carlsbad, CA), 1 × HEPES buffer (Cellgro, Manassas, VA), 1 × nonessential amino acids (Gibco), and penicillin/streptomycin/glutamine (Invitrogen) at a concentration of 2.5 × 105–2.5 × 106 cells ml−1. In a 96-well U-bottom tissue culture plate, 10 ng ml−1 recombinant human (rh)IL-2 (R&D Sytems, Minneapolis, MN), 10 ng ml−1 rhIL-6 (eBioscience), 10 ng ml−1 rhIL-1b (eBioscience), 10 ng ml−1 rhIL-23 (R&D Sytems), and anti-CD3/anti-CD28 activation beads (Miltenyi Biotec, Bergisch Gladbach, Germany) at a ratio of one bead per cell were added to each well. A decreasing concentration of rhTGF-β (R&D Sytems) was added to a series of five wells: 1, 0.1, 0.01, 0.001, and 0 ng ml−1, and 10 μg ml−1 anti-TGF-β antibody (R&D Systems) was added to a sixth well. After 3 days, the cells were split and 10 ng ml−1 rhIL-2 (R&D Sytems) was added to each well. On day 6, the cells were harvested, washed in 1 × PBS, and then stimulated for 4 h in PMA/Ionomycin for FACS analysis.
Hierarchical clustering. Unsupervised hierarchical clustering was performed using Cluster (Eisenlab, Berkeley, CA) on the 232 boolean gates generated from all two, three, four, and five possible combinations of IFNγ, TNFα, IL-17A, IL-22, and IL-4. Each row represents a different boolean gate combination, and each column represents a different biopsy sample. Yellow represents a greater expression than the mean of a particular boolean gate. Blue represents a lower expression than the mean, and black represents median expression.
Quantitative PCR. RNA was isolated from intestinal pinch biopsies using the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions. In all, 100 ng to 1 μg of RNA from each sample was reverse-transcribed to synthesize cDNA using Superscript III (Invitrogen). The resulting cDNA was used for quantitative real-time PCR using Taqman probes for TGF-β, c-Maf, IL13RA2, MMP7, CHI3L1, FoxP3, IL-17A, IL-6, IL-23, and IL-1b. All cycle threshold values were normalized to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) values and log transformed.
Statistical analysis. Statistical analysis was performed using Prism 5.0 (GraphPad Software, La Jolla, CA). The unpaired t-test was used to assess statistical significance for all the samples.
References
Strober, W. & Fuss, I.J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140, 1756–1767 (2011).
Kaser, A., Zeissig, S. & Blumberg, R.S. Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573–621 (2010).
Mayer, L. Evolving paradigms in the pathogenesis of IBD. J. Gastroenterol. 45, 9–16 (2010).
Maloy, K.J. & Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011).
Littman, D.R. & Rudensky, A.Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Brand, S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut 58, 1152–1167 (2009).
Trifari, S. & Spits, H. IL-22-producing CD4+ T cells: middle-men between the immune system and its environment. Eur. J. Immunol 40, 2369–2371 (2010).
Ouyang, W., Kolls, J.K. & Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467 (2008).
Sonnenberg, G.F., Fouser, L.A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Spits, H. & Di Santo, J.P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Colonna, M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity 31, 15–23 (2009).
Broadhurst, M.J. et al. IL-22+ CD4+ T cells are associated with therapeutic trichuris trichiura infection in an ulcerative colitis patient. Sci. Transl. Med. 2, 60ra88 (2010).
Achyut, B.R. & Yang, L. Transforming growth factor-beta in the gastrointestinal and hepatic tumor microenvironment. Gastroenterology 141, 1167–1178 (2011).
Rutz, S. et al. Transcription factor c-Maf mediates the TGF-beta-dependent suppression of IL-22 production in T(H)17 cells. Nat. Immunol. 12, 1238–1245 (2011).
Abraham, C. & Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 (2009).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Geboes, K. & De Hertogh, G. Indeterminate colitis. Inflamm. Bowel Dis. 9, 324–331 (2003).
Duhen, T., Geiger, R., Jarrossay, D., Lanzavecchia, A. & Sallusto, F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol 10, 857–863 (2009).
Volpe, E. et al. Multiparametric analysis of cytokine-driven human Th17 differentiation reveals a differential regulation of IL-17 and IL-22 production. Blood 114, 3610–3614 (2009).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Aujla, S.J. et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14, 275–281 (2008).
Abraham, C. & Medzhitov, R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140, 1729–1737 (2011).
Caporaso, J.G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Kim, C.J. et al. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 5, 670–680 (2012).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009).
Sugimoto, K. et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 (2008).
Zenewicz, L.A., Yancopoulos, G.D., Valenzuela, D.M., Murphy, A.J., Stevens, S. & Flavell, R.A. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29, 947–957 (2008).
Geremia, A. et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208, 1127–1133 (2011).
Sekikawa, A. et al. Involvement of the IL-22/REG Ialpha axis in ulcerative colitis. Lab. Invest. 90, 496–505 (2010).
Trifari, S., Kaplan, C.D., Tran, E.H., Crellin, N.K. & Spits, H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 10, 864–871 (2009).
Basu, R. et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075 (2012).
Rifkin, D.B. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J. Biol. Chem. 280, 7409–7412 (2005).
Monteleone, G., Boirivant, M., Pallone, F. & MacDonald, T.T. TGF-beta1 and Smad7 in the regulation of IBD. Mucosal Immunol. 1 (Suppl 1), S50–S53 (2008).
Clemente, J.C., Ursell, L.K., Parfrey, L.W. & Knight, R. The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012).
Cho, I. & Blaser, M.J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Caporaso, J.G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108 (Suppl 1), 4516–4522 (2011).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Blankenberg, D. et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19 Unit 19.10 1–21 (2010).
Acknowledgements
PL is supported by grants from the National Institutes of Health (AI093811 and AI094166), the Broad Medical Research Program, and the Kevin & Marsha Keating Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Leung, J., Davenport, M., Wolff, M. et al. IL-22-producing CD4+ cells are depleted in actively inflamed colitis tissue. Mucosal Immunol 7, 124–133 (2014). https://doi.org/10.1038/mi.2013.31
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2013.31
This article is cited by
-
Location is important: differentiation between ileal and colonic Crohn’s disease
Nature Reviews Gastroenterology & Hepatology (2021)
-
Immunomodulatory role of Parkinson’s disease 7 in inflammatory bowel disease
Scientific Reports (2021)
-
Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota
Nature Medicine (2020)
-
Targeting immune cell circuits and trafficking in inflammatory bowel disease
Nature Immunology (2019)
-
JAK inhibition as a therapeutic strategy for immune and inflammatory diseases
Nature Reviews Drug Discovery (2017)