Abstract
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A)/TNF superfamily member 15 (TNFSF15) is a proinflammatory cytokine and TNFα superfamily member that is linked preclinically and clinically to inflammatory bowel disease (IBD). By homology and function, TNFα is its closest family member. In this study, we investigated the mechanism of TL1A-induced inflammation in CD4+ T cells and compared it with the TNFα pathway. We found that TL1A induces proinflammatory cytokines, including TNFα, from isolated human CD4+CD161+ T cells, whereas these cells were resistant to TNFα treatment. Anti-TNFα failed to block TL1A-induced cytokine production, indicating that the effects of TL1A are direct. Lastly, CD161 and TL1A expression were significantly and selectively increased in gut tissue biopsies, but not in the peripheral blood, from IBD patients. Thus, TLIA not only functions upstream of TNFα, driving its expression from CD161+ T cells, but is also independent of TNFα. These findings may have therapeutic IBD implications.
Similar content being viewed by others
Introduction
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A)/TNF superfamily member 15 (TNFSF15) is a proinflammatory cytokine and a member of the TNFα superfamily, as it was originally characterized in a screen for novel molecules with homology to TNFα.1, 2 TL1A is expressed on activated T cells, dendritic cells, and monocytes, as well as induced by TNFα and interleukin (IL)1α in Huvec cells in vitro.1, 3, 4, 5 In contrast to the TNFα receptors, which are expressed on essentially all cells, the receptor for TL1A, death receptor 3 (DR3), is primarily expressed on T cells, natural killer (NK) cells, and NK T (NKT) cells, thereby limiting the effects of TL1A.1, 3 A nonfunctional, soluble receptor, decoy receptor 3 (DcR3), also binds to but inhibits TL1A activity.1 In the presence of T-cell receptor stimulation, TL1A potentiates T-cell activation, cytokine production, and proliferation.1, 2, 3
Owing to its proinflammatory and costimulatory activity, TL1A has been linked to clinically active inflammatory bowel disease (IBD).6, 7 Genome-wide association study studies have identified polymorphisms in the TL1A gene that lead to increased susceptibility to IBD in multiple ethnic populations.8, 9, 10, 11, 12 Moreover, several studies have demonstrated an increase in TL1A RNA in tissue biopsies, as well as DR3 expression on CD4+ T cells in IBD patients.13, 14, 15 In addition, previous data have demonstrated that ulcerative colitis (UC) patients with both active and quiescent disease and Crohn’s disease (CD) patients with colon involvement have slightly elevated serum TL1A levels.16, 17
It is well established that TNFα plays a significant role in IBD, Since, neutralizing TNFα therapies are the first-line biological therapy, following immunosuppressants and steroids.7, 18, 19 While 60% of CD patients respond to anti-TNFα therapies, 40% remain resistant and at least half of the initial anti-TNFα responders become refractory to anti-TNFα within a year.20 Therefore, there is significant room for improvement in IBD therapies.
Similar to TNFα, TL1A is a trimeric molecule that is expressed as a membrane protein as well as a proteolytically cleaved soluble form.21 However, very little data are available regarding membrane-only TL1A, as most studies have solely investigated the function of soluble TL1A. For instance, in the presence of both IL12 and IL18, soluble TL1A can significantly enhance interferon (IFN)γ expression in CD4+ T cells.22, 23 As both IL12 and IL18 are upregulated in CD patients and T-helper (Th)1 cytokines have long been implicated in CD pathology, TL1A may exacerbate disease via potentiating proinflammatory cytokine production.24, 25, 26, 27
CD161 (also known as KLRB1) is an NK-like receptor expressed on all NK cells, and is also expressed on approximately 25% of blood-resident CD4+ T cells. While 50–60% of lymphocytes cells in the gut are CD3+CD4+CD161+, the number of CD4+CD161+ T cells in CD patient lesions is increased, suggesting that CD161+ T cells have a critical role in CD pathogenesis.28, 29, 30 CD4+CD161+ T cells have a Th17-like phenotype, producing IL17, while also producing IFNγ.30 Even though CD4+CCR6+CD161+ T cells are Th17 in origin, at sites of inflammation, they revert into a Th1-like phenotype.31 A recent report demonstrated an increase in IFNγ production in blood-derived CD4+CD56−CD161+ T cells following agonistic anti-DR3 stimulation.4 As TL1A has a propensity to drive inflammation in the gut, this cell population may have a unique role in gut-specific TL1A biology.
In this study, we investigated the mechanism of TL1A-induced inflammation in human CD4+ T cells and compared the effects to the TNFα-induced pathway. Both soluble and membrane TL1A directly induces proinflammatory cytokines from CD4+CD161+ T cells, while these cells are resistant to TNFα. More importantly, TL1A induces TNFα expression from CD161+ T cells, suggesting that TL1A is upstream of TNFα. Soluble and membrane TL1A-induced cytokine expression was inhibited by a novel neutralizing anti-TL1A antibody, but not by an anti-TNFα antibody, indicating that TL1A directly affects CD161+ T cells. Finally, we have demonstrated that CD161 and TL1A expressions are both significantly elevated in gut biopsies, but not in the peripheral blood, of CD patients. We propose that TL1A drives gut inflammation both in parallel and upstream of TNFα, via direct potentiation of TNFα and other proinflammatory cytokines from CD4+CD161+ T cells. Thus, neutralization of TL1A has the potential to block two pathways important to the pathophysiology of IBD.
Results
Soluble TL1A directly induces TNFα, IFNγ, and IL2 expression from anti-CD3-activated CD4+ T cells
To determine the effects of TL1A on human CD4+ T cells, anti-CD3-activated T cells were treated with TL1A and a panel of selected cytokines and activation markers were analyzed by Taqman, enzyme-linked immunosorbent assay, and flow cytometry. By quantitative polymerase chain reaction (qPCR), we found that soluble TL1A induced expression of TNFα, IL2, IFNγ, and CD25 that was inhibited by soluble DcR3-Fc (Figure 1; P<0.01). In contrast, TL1A was unable to potentiate the expression of cytotoxic T-lymphocyte antigen 4 (CTLA4) or TNF receptor II (TNFRII). While IL2, IFNγ, and CD25 were all predicted as responsive to TL1A, we were surprised to see that TL1A induced TNFα from CD4+ T cells.1 We confirmed TL1A-induced protein expression of TNFα over time (Figure 1b). In addition, we demonstrated an increase in IL2, IFNγ, and CD25 expression, as well as a slight increase in DR3 expression over time (Supplementary Figure 1 online). As TNFα also has direct effects on CD4+ T-cell activation and proliferation, we treated cells with TL1A plus a neutralizing anti-TNFα antibody and demonstrated that IL2 expression was not affected (Figure 1c). Thus, the effects of TL1A on CD4+ T cells are direct and not secondary owing to TNFα expression.
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A) induces proinflammatory cytokine production, including TNFα, from CD4+ T cells. (a) Naïve human CD4+ T cells were activated with anti-CD3 plus TL1A, in the presence or absence of 10 μg ml−1 decoy receptor 3 (DcR3)-Fc inhibitor. After 48 h, RNA was harvested and gene expression analyzed via quantitative polymerase chain reaction (qPCR). (b) Naïve CD4+ T cells were treated as in a, but supernatants were harvested at 24, 48, or 72 h and TNFα was measured by AlphaLISA. (c) Naïve human CD4+ T cells were treated as in a, but cells were also treated with 10 μg ml−1 neutralizing anti-TNFα antibody or isotype control, for 48 h. Interleukin (IL)-2 was measured by AlphaLISA; n=4 replicates per group. CTLA4, cytotoxic T-lymphocyte antigen 4; hTNF, human TNF; IFN, interferon; NT, not receiving any treatment; TNFRII, TNF receptor II.
Neutralizing anti-human TL1A antibody-0011 potently inhibits both soluble and membrane-bound TL1A
To investigate the role of TL1A in CD4+ T-cell biology, we generated a mouse anti-human TL1A monoclonal antibody-0011. To confirm its neutralizing activity, CD4+ T cells were treated with soluble TL1A plus increasing concentrations of anti-TL1A-0011 for 24 h and found that anti-TL1A-0011 neutralized soluble TL1A-induced IL2 production from anti-CD3-activated human CD4+ T cells (Figure 2a; P⩽0.05). As TL1A-induced phosphorylation of nuclear factor-κB is part of the TL1A signaling pathway, we measured phosphorylation of nuclear factor-κB in CD4+ T cells by intracellular phospho-flow cytometry.32 TL1A induced phospho-nuclear factor-κB in CD4+ T cells within 10 min, which can be completely inhibited by anti-TL1A-0011, in a dose-dependent manner (Figure 2b,c; P<0.005).
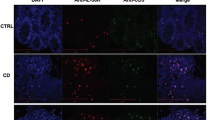
Neutralizing antitumor necrosis factor-like cytokine 1A (TL1A) antibody-0011 inhibits both soluble (s) and membrane-bound TL1A (MbTL1A). (a) Human CD4+ T cells were treated as in Figure 1c, but increasing amounts of anti-TL1A-0011 was added to the culture before interleukin (IL)2 analysis; n=3 replicates per group. (b) Human peripheral blood mononuclear cells (hPBMCs) were treated with TL1A plus increasing concentrations of anti-TL1A-0011 or isotype (iso) control for 10 min. Cells were gated on CD4+ and phosphorylation of nuclear factor-κB (ph-NF-κB) was measured by flow cytometry. (c) Purified human CD14+ monocytes were activated with immune complexes and then stained with mouse anti-human TL1A-0011 antibody or immunoglobulin G (IgG)2A isotype control antibodies (original magnification: 400 × ). (d) Empty Chinese hamster ovary (CHO) cells or MbTL1A-expressing CHO cells were stained with anti-TL1A-0011 and analyzed via immunofluorescence. Full-length TL1A and MbTL1A were expressed in CHO cells and supernatants were analyzed via TL1A enzyme-linked immunosorbent assay (ELISA) (Enzo) (original magnification: 525 × ). (e) MbTL1A-CHO cells were stained with recombinant human (rh) DR3-Fc or decoy receptor 3 (DcR3)-Fc, and anti-Fc-fluorescein isothiocyanate (FITC)-conjugated secondary antibody was used to detect expression. (f) Human CD4+ T cells were cultured on anti-CD3-coated plates plus empty CHO cells, CHO-MbTL1A cells, or 200 ng ml−1 sTL1A for 24 h. In some cases, cells were also treated with 10 μg ml−1 anti-TL1A-0011. After 24 h, supernatants were harvested and cytokine measured by Luminex. Samples were run in duplicate and a minimum of three times. DR3, death domain-containing receptor; FL, Fas ligand; NT, not receiving any treatment.
Although TL1A is functional in both soluble and membrane-bound (Mb) forms, comparison between the two has thus far been restricted to induction of IFNγ expression.21 Therefore, we next tested whether anti-TL1A-0011 could bind to and inhibit soluble and membrane TL1A. Using immunofluorescence, we confirmed that anti-TL1A-0011 binds to endogenous membrane TL1A on immune complex-activated monocytes (Figure 2c). Owing to the low percentage of cells expressing membrane TL1A (⩽20%), we generated a noncleavable, membrane-only TL1A Chinese hamster ovary (CHO) cell line (MbTL1A) by replacing amino acids 67–94 in full-length human TL1A with a flexible (G4S)4 linker.33 Compared with transfection of full-length TL1A into CHO cells, which express both soluble and membrane TL1A, MbTL1A-CHO clones expressed only membrane TL1A, but not soluble TL1A (Figure 2d). In addition, flow cytometry confirmed that DR3Fc, DcR3-Fc, as well as anti-TL1A-0011 bind to MbTL1A, indicating that we have maintained receptor binding (Figure 2e). To compare the biology of the two forms of TL1A, purified human CD4+ T cells were cocultured with fixed amounts of MbTL1A or soluble TL1A plus anti-CD3 and cytokine production was measured after 24 h. We found that MbTL1A and soluble TL1A were capable of potentiating TNFα, IL2, and IFNγ production from CD4+ T cells (Figure 2f; P⩽0.04). In addition, expression of granulocyte–macrophage-colony-stimulating factor, IL4, and IL5 was increased (Supplementary Figure 2 online). Other than IFNγ, whose expression was only partially blocked by anti-TL1A-0011, MbTL1A-induced expression of the remaining cytokines is completely blocked by anti-TL1A-0011. Thus, soluble and membrane TL1A are both capable of inducing TNFα in anti-CD3-activated CD4+ T cells, which can be inhibited by anti-TL1A-0011.
Both soluble and MbTL1A directly potentiate IFNγ and TNFα expression from IL12- and IL18-stimulated CD3+ T cells
Previous reports demonstrated that TL1A synergizes with IL12 and IL18 to potentiate IFNγ expression in CD4+ T cells.18, 22, 23 As IL12 and IL18 are both expressed and create a proinflammatory environment in CD patients, we used these stimulation conditions to analyze the effects of TL1A compared with TNFα. We found that both cytokines induced IFNγ in a dose-dependent manner (Figure 3a) and anti-TL1A-0011 inhibited TL1A-induced IFNγ expression (Figure 3b; P⩽0.004). Interestingly, TL1A was again able to induce low levels of TNFα in whole blood (P⩽0.03) and treatment with anti-TNFα did not significantly inhibit soluble TL1A-induced IFNγ expression (Figure 3c,d). TL1A also induced TNFα from isolated human peripheral blood mononuclear cells (hPBMCs), which was inhibited with anti-TL1A-0011 (Figure 3e; P=3 × 10−6). Thus, TNFα is also induced by TL1A with IL12 plus IL18 treatment, indicating that it is a reproducible response, irrespective of the stimulation conditions.
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A) directly induces TNFα and interferon (IFN)γ in interleukin (IL)12 plus IL18-stimulated whole blood. (a) Peripheral blood was treated with IL12 and IL18 plus increasing concentrations of TL1A or TNFα for 24 h. Supernatants were analyzed for IFNγ expression by AlphaLISA. (b) Whole blood was treated with 50 ng ml−1 TL1A as in a, but increasing concentrations of anti-TL1A-0011 antibody was added. IFNγ was measured as above. (c) Whole blood was treated as in a, but TNF-α was analyzed instead by AlphaLISA. (d) Whole blood was treated with 50 ng ml−1 of TL1A or TNFα, plus 15 μg ml−1 anti-TL1A-0011 or anti-TNFα, and IFNγ was analyzed. (e) Human peripheral blood mononuclear cells (hPBMCs) were treated with IL12 and IL18 plus TL1A, anti-TL1A-0011, or isotype (iso) control, and the total amount of TNFα was measured in the supernatant after 24 h. (f) hPBMCs were cultured with IL12, IL18, and increasing amounts of membrane-bound TL1A (MbTL1A) Chinese hamster ovary (CHO) cells and IFNγ was measured. Untransfected CHO cells did not induce IFNγ (data not shown). (g) A total of 5 × 104 CHO-MbTL1A cells were cultured with hPBMCs, IL12, IL18, plus increasing concentrations of anti-TL1A-0011, anti-TNFα, or iso control. IFN-γ was measured as above after 24 h. (h, i) hPBMCs were treated with IL12 and IL18 plus 50 ng ml−1 TL1A. Cells were then gated on CD3+ T cells, CD3+CD56− natural killer (NK) cells, CD3+CD56+ NK T (NKT) cells, or CD3−CD56− lymphocytes, and (g) the percentage of IFNγ-producing cells or (h) the mean fluorescence intensity (MFI) of IFNγ was analyzed by flow cytometry. All samples were run with a minimum of three replicates per sample. IgG, immunoglobulin G; NT, not receiving any treatment.
Repeating the IL12–IL18 assay with MbTL1A cells, we found that there was a dose-dependent increase in IFNγ production, which could be inhibited by anti-TL1A-0011 (Figure 3f,g; P=0.0006). Even though TNFα was induced by MbTL1A in CD4+ T cells, anti-TNFα was unable to significantly block the MbTL1A-induced IFNγ expression (Figure 3g). Other cytokines induced by both MbTL1A and soluble TL1A, but not TNFα in hPBMCs, are granulocyte–macrophage colony-stimulating factor, IL4, IL5, and IL13 (Supplementary Figure 3 online). Thus, TL1A directly induces IFNγ and TNFα from both whole blood and hPBMCs, independent of TNFα expression.
As both T lymphocytes and NK cells express DR3, we next investigated which cell population was responding to TL1A. Using flow cytometry, hPBMCs were subdivided into CD3+CD56− T cells, CD3−CD56+ NK cells, and CD3+CD56+ NKT cells. In the presence of IL12 and IL18, NK cells, NKT cells, and CD3+ T cells increased the amount of IFNγ produced per cell (Figure 3h). Although a significant proportion of T cells and NKT cells produce IFNγ, only CD3+ T cells increased the number of cells producing IFNγ following TL1A treatment (Figure 3i; 1.8-fold increase compared with NK and NKT cells). TNFα induced IFNγ expression in NK cells (data not shown).
TL1A drives IFNγ expression in CD3+CD56−CD161+ T cells, while they remain resistant to TNFα
When hPBMCs were treated with IL12 and IL18 plus an agonistic anti-DR3 antibody, the percentage of IFNγ-producing CD161+ T cells increased.18 However, before this study, (1) it was unknown whether these cells also respond to TNFα and (2) if so, whether TL1A differentially increases the expression of other cytokines in CD161+ T cells, in addition to IFNγ. As reported previously, TL1A increases IFNγ expression in CD161+ T cells, whereas CD161− cells do not respond (Figure 4a). Remarkably, however, when cells were treated with TNFα, we found that, unlike TL1A, TNFα does not induce IFNγ expression in CD3+CD56−CD161+ cells, despite inducing IFNγ in the whole blood assay (Figure 4b). In contrast to TNFα, TL1A consistently increases the percentage of CD161+ T cells that respond, as well as the amount of IFNγ produced per cell (Figure 4c,d; P=0.009). TL1A induces an average 3.2-fold increase in IFNγ expression, whereas TNFα induces only an average 1.5-fold increase (P=0.008), across eight different donors. Consistent with this, IL12 and IL18 treatment increased DR3 levels solely on CD161+ cells, but not CD161− cells (Supplementary Figure 4A online). However, the expression of TNFRII was also higher on CD161+ T cells compared with CD161− T cells (Supplementary Figure 4B online). Expression of TNFRI was higher in CD161− cells, but the overall levels were very low (mean fluorescence intensity <30; data not shown). As expected, levels of the chemokine receptor, CCR6, were higher on CD161+ T cells (Supplementary Figure 4C online). Thus, TNFα nonresponsiveness in CD161+ T cells is not due to absent receptor expression levels. Finally, we demonstrated that anti-TL1A-0011 inhibited TL1A-induced IFNγ production from CD161+ T cells, whereas anti-TNFα was unable to inhibit significantly the amount of IFNγ produced (Figure 4e,f). In this donor, anti-TNFα modestly decreased the number of IFNγ-producing CD161+ cells. Overall, TL1A directly affects CD161+ T cells.
Unlike tumor necrosis factor (TNF)α, TNF-like cytokine 1A (TL1A) directly induces interferon (IFN)γ expression in CD3+CD56−CD161+ T cells. (a) Human peripheral blood mononuclear cells (hPBMCs) were left untreated, treated with TL1A, interleukin (IL)12, and IL18, or IL12 and IL18 plus 200 ng ml−1 TL1A for 24 h. Cells were gated on CD3+CD56 and separated into (top panel) CD161+ or (bottom panel) CD161−, and intracellular IFNγ was measured by flow cytometry. (b) hPBMCs were treated as in a, but in this case 200 ng ml−1 TNFα was also added. (c, d) hPBMCs were harvested from eight healthy volunteers and each was treated with IL12 and IL18 plus either TL1A or TNFα. (c) The percent IFNγ-positive cells and (d) the mean fluorescence intensity (MFI) of IFNγ were analyzed by flow cytometry. (e, f) hPBMCs were treated as in a, but in certain cases, anti-TL1A-0011 or anti-TNFα, plus respective isotype (iso) controls, were added to the culture. After 24 h, (e) the percentage of IFNγ CD161+ cells or (f) the MFI of IFNγ was analyzed. Each sample was run in quadruplicate. NS, not significant; NT, not receiving any treatment.
TL1A, but not TNFα, induces proinflammatory cytokine expression, including TNFα, IFNγ, IL6, and IL17, from isolated CD4+CD161+ T cells
Although we confirmed that TL1A does not rely on TNFα to induce proinflammatory cytokines from CD161+ T cells, other cytokines may be induced in CD161− T cells that indirectly affect CD161+ T cells. Therefore, we isolated CD161+ or CD161− cells and analyzed TL1A- or TNFα-induced expression of a broad array of proinflammatory cytokines. We initially analyzed intracellular IFNγ and TNFα expression and found that CD161+ T cells produced both cytokines in response to TL1A, but not to TNFα (Figure 5a–c; P⩽0.01). Analysis of the supernatants confirmed induction of both IFNγ and TNFα by TL1A in CD161+ T cells (Figure 5b,d). In general, CD161− T cells did not respond to TL1A, but there was a slight increase in IFNγ production, although the amount of IFNγ produced by CD161+ is significantly different from CD161− cells (Figure 5c; P=0.006). We next analyzed the supernatants for the expression of a 17-plex panel of cytokines. With the exception of IL8, which is expressed by both CD161+ and CD161− cells, TL1A induces significant levels of IL6, IL17, granulocyte–macrophage colony-stimulating factor, and IL13 from CD161+ compared with CD161− T cells (Figure 5g; P⩽0.0002). The remaining cytokines analyzed in the multiplex enzyme-linked immunosorbent assay kit were either not induced by IL12 and IL18 or not potentiated by TL1A (data not shown). TNFα did not induce cytokine expression in either CD161+ or CD161− cells, again except for IL8. Therefore, other than IL8, TL1A induces an array of proinflammatory cytokines primarily from CD161+ T cells, which TNFα is unable to do. Depletion of CD4+ CD161+ T cells may be sufficient to eliminate TL1A-induced proinflammatory cytokine expression.
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A), but not TNFα, induces proinflammatory cytokine expression in isolated CD4+CD161+ cells, but not CD4+CD161− cells. (a–e) Human CD4+ T cells were purified and sorted into CD161+ and CD161− populations. Isolated cells were treated with IL12 and IL18 plus either TL1A or TNFα for 36 h. (a) Intracellular interferon (IFN)γ or (c) TNFα was measured by flow cytometry, while supernatants were analyzed for (b) IFNγ or (d) TNFα expression. (e) Cytokine expression was measured by 17-plex Luminex kits. Each sample had six replicates. GM-CSF, granulocyte–macrophage colony-stimulating factor; NS, not significant; NT, not receiving any treatment.
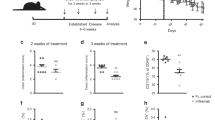
To investigate whether TL1A is indeed upstream of TNFα in an in vivo setting, we compared the effects of a surrogate antimouse TL1A and antimouse TNFα antibodies in an acute 2,4,6-trinitrobenzene sulfonic acid-induced model of colitis. Mice treated with either anti-TL1A or anti-TNFα showed comparable and significant inhibition of disease progression, as measured by clinical score compared with isotype control-treated animal, as expected (Supplementary Figure 5A online and Meylan et al.34). When baseline and terminal serum samples were analyzed for cytokine expression, TNFα expression was significantly inhibited by anti-TL1A, as predicted by our in vitro studies (Supplementary Figure 5C online). Thus, TL1A appears to be upstream of TNFα in an IL12-dependent in vivo colitis model. Future studies should address the role of TL1A in chronic models of colitis (i.e., CD45RBhi T-cell transfer model) and the efficacy of anti-TL1A and anti-TNFα as a combination therapy in vivo.
Serum levels of TL1A and responsiveness of blood-derived CD161+ T cells in CD patients are similar to healthy volunteers
We next analyzed the responsiveness of CD3+CD56−CD161+ T cells derived from the blood of CD patients compared with healthy volunteers (HVs). Following treatment with TL1A, there was no difference in the average percentages of CD161+ T cells in HVs vs. CD patient samples and treatment with IL12/IL18 and/or TL1A did not affect the number of CD161+ T cells (Figure 6a). Similar to HV-derived CD161+ T cells, CD patient-derived CD161+ T cells treated with TL1A still induced IFNγ expression, whereas TNFα did not (Figure 6b). However, there was no difference between the magnitude of the response between HVs (Supplementary Figure 6 online and Figure 3) and CD patient-derived CD161+ T cells (Figure 6b), indicating that CD patient-derived CD161+ T cells from the peripheral blood are not more responsive to TL1A. While the levels were low, there was no difference in DR3 levels (data not shown).
Peripheral blood-derived CD161+ T cells from Crohn’s (CD) patients are not more responsive to tumor necrosis factor (TNF)-like cytokine 1A (TL1A) and do not have elevated serum TL1A levels. (a, b) Peripheral blood mononuclear cells (PBMCs) were purified from peripheral blood of healthy volunteers (HVs) or CD patients (n=3 each) and treated with interleukin (IL)12 and IL18 plus TL1A or TNFα, similar to Figure 4. (a) The percentage of CD3+CD56−CD161+ T cells were analyzed by flow cytometry. (b) Interferon (IFN)γ mean fluorescence intensity (MFI) was measured after 24 h in CD patients. Patient characteristics are listed in Table 1. (c, d) Serum samples were collected from control, ulcerative colitis (UC), and CD patients are analyzed for the expression of (c) TL1A (c) or (d) TNFα. Each patient or HV CD161+ sample was run in triplicate and then averaged. For serum samples, each patient serum was analyzed in duplicate and averaged. NT, not receiving any treatment.
Although they are not more responsive, CD patients may have elevated serum levels of TL1A, leading to increased expression of serum IFNγ or other proinflammatory cytokines. However, when we analyzed serum cytokine coexpression, neither TL1A nor TNFα levels were elevated in CD or UC patient samples compared with controls (Figure 6c,d). Thus, TL1A expression and CD161+ T-cell responses are comparable between CD patients and HVs.
Both TL1A and CD161 are significantly upregulated in CD patient biopsies
While there is limited biological expression of TL1A in the peripheral blood of IBD patients, we next investigated the levels of TL1A, TNF-α, and CD161 in gut biopsies taken from the same patients described in Figure 6c,d. Control, UC patient, and CD patients, which were further subdivided into disease localized in either the ileum or the colon, were analyzed for the expression of a panel of TL1A-associated genes. Only those genes whose expression was significantly increased with a P<0.05 and a fold change (FC) >1.5 were scored as significantly different (*). Microarray analysis demonstrated that TL1A was elevated in UC patients (P=0.0005; FC=1.9), ileum of CD patients (P=0.0000003; FC=2.4), and colon of CD patients (P=0.02; FC=1.5), whereas CD161/KLRB1 was significantly and specifically increased only in the ileum of CD patients (P=0.00006; FC=1.9). In UC patients, TNFα and TL1A were both increased (Figure 7a). While TNFα was also significantly increased in the colon (P=0.03) and ileum (P=0.007) of CD patients, the fold induction was only 1.4; therefore, based on our criteria, TNFα levels in CD patients did not score as significantly different from controls. IFNγ and DcR3/TNFRSF6B expression were significantly increased in UC and CD patients, whereas there was no change in IL17A, or DR3 expression (Figure 7a,b). CXCL10 expression was increased in all patient samples, indicating that all patients had active disease (Figure 7b).
Tumor necrosis factor (TNF)-like cytokine 1A (TL1A) and CD161 are both significantly and selectively upregulated in the ileum of Crohn’s disease (CD) patient biopsies. Biopsies were harvested from control patients (n=22), ulcerative colitis (UC) patients (n=17) from CD patients with disease in the colon, and (n=23) CD patients with disease in the ileum (n=25), and RNA was analyzed via microarray. (a) Expression of TL1A, CD161, and TNFα and DR3 or (b) decoy receptor 3 (DcR3), interferon (IFN)γ, interleukin (IL)17A, and CXCL10 were normalized via the robust multiarray average (RMA) algorithm. P-values were determined by standard t-test. Patient characteristics are listed in Table 2. DR3, death domain-containing receptor; TNFSF15, TNF superfamily member 15; TNFRSF25, TNF receptor superfamily member 25.
Previous studies have suggested that TNFα can induce TL1A expression in Huvec cells and treatment of rheumatoid arthritis patients with TNFα inhibitors led to a decrease in serum TL1A levels.1, 16 To determine what effect anti-TNFα treatment has on TL1A gene expression, we segregated total CD patient samples into (1) patients with no treatment (NT), (2) patients undergoing anti-TNFα treatment, and (3) patients being treated with first-line, standard of care (SOC). We found that in CD patients, TL1A levels decreased with anti-TNFα or SOC treatment, compared with NT samples, but the levels did not go back to control (Figure 8a). Moreover, there was no difference in the levels of TL1A between patients treated with anti-TNFα or SOC. Interestingly, TNFα RNA levels did not decrease following anti-TNFα or SOC treatment (Figure 8b). In addition, levels of CD161 and IFNγ remained the same whether patients were not treated, or underwent anti-TNFα or SOC treatment (Figure 8c).
Crohn’s disease patients treated with anti TNFα or standard of care (SOC) have reduced expression levels of TL1A, compared to CD patients without treatment. CD patient samples from Figure 7 were segregated into patients not receiving any treatment (NT, n=12), treated with anti-TNFα therapies (n=10), or SOC, including 5-ASA, azathioprine, methylprednisilone, methotrexate and steroids (n=22) and compared to control, non-IBD patient biopsies (n=22). Expression of TL1A (c), TNFα (d) or CD161 and IFNγ (e) were analyzed by microarray as in Figure 7 and P-values were generated via student's t-test. NS, non-significant.
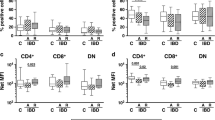
As we were unable to confirm the expression of TL1A pathway genes as well as several genes associated with TL1A biology in these patient samples, we collected a second set of active CD and non-IBD, non-irritable bowel syndrome (IBS) control patient samples and measured gene sets by qPCR. CD161, TNFα, IFNγ, and DcR3 are all significantly upregulated in CD patient biopsies compared with control samples (Figure 9a,b; P<0.001). In addition, levels of IL18 were significantly elevated, whereas IL12 was increased but was not significant (Figure 9b; P<0.01). In contrast, levels of DR3 and IL17 were again not significantly elevated (Figure 9b,c). Finally, levels of the homing receptors, CCR6, integrin-α4, and -β7, were all significantly elevated, while CCR9 was increased but not significantly (Supplementary Figure 7 online). Finally, similar to microarray data, CD4 levels were not increased, while CXCL10 was. We also were able to segregate patients based on anti-TNFα treatment; however, all patients were being treated so that there were no untreated controls. In these samples, there was no difference in TL1A, TNFα, or CD161 level in patients treated with anti-TNFα and other medications, similar to our microarray data (Supplementary Figure 8 online).
Expression of tumor necrosis factor (TNF)-like cytokine 1A (TL1A) and CD161 is confirmed in quantitative polymerase chain reaction (qPCR) Crohn’s disease (CD) patient biopsies. Biopsies were obtained from 24 noninflammatory bowel disease (IBD), nonirritable bowel syndrome (IBS) controls, or 13 CD patients with active disease. RNA was harvested and the following genes were analyzed by qPCR: (a) TL1A, TNFα, and CD161; (b) IFNγ, DR3, decoy receptor 3 (DcR3); and (c) IL17A, IL12a, and IL18. Patient characteristics are listed in Table 3. DR3, death domain-containing receptor. ***P<0.001.
Discussion
Dysregulation of intestinal CD4+ T subsets is implicated in IBD etiology, with over exuberant production of Th1 (IFNγ and TNFα) and Th17 (IL17) cytokines predominating in CD, and Th2 cytokines (e.g., IL13) and IL17 predominating in UC.35 TL1A is a proinflammatory and T-cell costimulatory cytokine, which is active in both soluble and membrane-bound forms. TL1A promotes both Th1 and Th17 responses, making it a very attractive IBD target.2 In addition, TL1A is a member of the TNFα superfamily, sharing overlapping attributes with its founding member, TNFα. In this study, we investigated the mechanism of TL1A-induced inflammation in CD4+ T cells, and compared these effects with TNFα, to determine whether the TL1A pathway is independent, overlapping, or partially convergent with the TNFα pathway. We found that both soluble and MbTL1A directly induces proinflammatory cytokines from CD161+ T cells, while these cells are resistant to TNFα. More importantly, TL1A induces TNFα solely from CD4+CD161+ T cells, a population of cells implicated in CD.30 In contrast, compared with CD161+ T cells, TL1A does not induce significant cytokine expression in CD161−T cells. While anti-TL1A-0011 inhibited both soluble and MbTL1A-induced proinflammatory cytokines, neutralizing anti-TNFα antibodies generally did not inhibit TL1A-induced cytokine expression, indicating that TL1A is directly driving cytokine expression. Taken together, our data demonstrate that TL1A is both upstream and independent of TNFα. As CD161+ T cells are highly prevalent in the gut tissue, TL1A-driven TNFα may be a critical local source of TNFα in IBD. TL1A can capitalize on the proinflammatory environment to potentiate IFNγ and TNFα expression in the gut, but does so independently of TNFα.
A previous study reported that anti-CD3−CD28 stimulation of CD4+CD161+ T cells produced high levels of IL17 but not IFNγ; IFNγ levels only increased when IL1β and IL23 were added.30 However, in our study, TL1A potentiated IL17 expression in isolated CD161+ T cells treated with IL12 and IL18. Previous data as well as our current qPCR data support a role for these cytokines in CD patients.24, 25Thus, different cytokine milieu and activation strategies are both critical for defining TL1A-induced T-cell responsiveness and downstream gene responses.
To provide clinical relevance to the TL1A− CD161+ T-cell pathway, peripheral blood-derived CD161+ T-cell responses, as well as serum levels of TL1A and TNFα with corresponding gene expression in gut biopsies were analyzed in IBD patient samples. First, CD161+ T cells derived from CD patients were not more responsive to TL1A compared with HVs. Second, using serum samples obtained from the same patient samples analyzed by microarray, we found that TL1A levels were not elevated in the peripheral blood of CD patients. Previous studies have shown that TL1A serum levels are slightly elevated in CD and UC patients.15, 17 However, when TL1A expression in IBD patient serum was compared with their corresponding gut biopsies, we found that TL1A appears to be associated with localized disease in the gut, but not in the peripheral blood, of IBD patients
While previous data also demonstrated elevated TL1A levels in CD and UC patients using qPCR, we have demonstrated elevated expression of the TL1A pathway and relevant associated genes, including CD161 and IFNγ, in IBD patients by microarray.13, 14, 15, 17 Moreover, using qPCR, the expression of TL1A and CD161 was confirmed in another set of CD patient samples with active disease. Drawing from our mechanistic in vitro data, high levels of TL1A may exacerbate disease via proinflammatory cytokine expression from CD161+ T cells localized in the gut. Compared with elevated levels in UC patients, TNFα levels in CD patient biopsies had a P-value of <0.05, but only had a fold increase of 1.4 by microarray. Thus, based on our criteria, TNFα was not significantly upregulated in CD patients. However, using qPCR, we demonstrated that TNFα was elevated in this CD patient subset. Compared with microarray technology, TNFα levels in qPCR samples may register as significantly increased owing to the qPCR amplification technique used to analyze gene expression.
Previous data in RA patient serum has demonstrated inhibition of TL1A by anti-TNFα.16 We found that anti-TNFα and SOC therapies partially inhibited TL1A levels, whereas other genes, including TNFα RNA levels itself, remained unaffected. If anti-TNFα or SOC treatment can control disease severity in certain patients, it stands to reason that levels of TL1A, if associated with disease progression, would also decrease. Future studies should directly analyze TL1A levels in patients before and following anti-TNFα treatment, and correlate it to disease severity vs. remission. To better tailor anti-TL1A therapies, the TNFα-responsive target cell producing TL1A in gut biopsies should also be investigated. For the first time, were analyzed TL1A plus associated genes in IBD patient biopsies, providing strong evidence of the relevance of the TL1A pathway, relative to TNFα expression in IBD.
While TNFα was previously shown to induce TL1A in endothelial cells, only now do we appreciate that TL1A drives TNFα expression, specifically from CD161+ T cells, demonstrating an intriguing positive feedback loop between the two cytokines.1 Using neutralizing anti-TNFα and anti-TL1A antibodies, we demonstrated that TL1A drives proinflammatory cytokine expression (i.e., IFNγ) independently of TNFα. Treatment with anti-TL1A antibody could therefore potentially inhibit both these pathways in patients. However, there are other sources of TNFα in the gut that do not respond to TL1A (i.e., macrophages) and will not respond to anti-TL1A treatment. In this scenario, anti-TNF plus anti-TL1A combination therapy may be beneficial, especially in those patients with high TNFα and TL1A levels. In addition, those CD patients who have become refractory to anti-TNFα therapies may use the TL1A pathway to bypass the need for TNFα to drive disease. Future studies should address whether the levels of TL1A are elevated in CD patients who are resistant to anti-TNFα therapies.
In conclusion, while there is some overlap between the two pathways (i.e., IFNγ production from peripheral blood), overall TL1A and TNFα target different cell types, and consequently, induce different proinflammatory pathways. Anti-TL1A therapy for CD patients could be beneficial for two reasons: (1) TL1A drives TNFα expression from CD161+ T cells, which would contribute to local disease exacerbation and (2) TL1A induces a proinflammatory pathway in CD161+ T cells that is independent of TNFα, but includes expression of IFNγ and other cytokines that could contribute to disease pathology.
Methods
Whole blood, hPBMCS, and CD4+ T cells assays. To obtain peripheral blood from HVs, Roche established an internal blood donation program, which was reviewed and received approval from an outside IRB (Copernicus Group IRB, Research Triangle Park, North Carolina). All volunteers were properly consented before becoming an active donor. For the whole blood assay, heparinized blood was treated with 2 ng ml−1 IL12 and 20 ng ml−1 IL18 (R&D Systems, Minneapolis, MN) and 50 ng ml−1 soluble TL1A for 24 h. Supernatants were assayed for IFNγ expression by AlphaLISA (Perkin-Elmer, Waltham, MA). Naïve CD4+ T cells were isolated from PBMC of HVs using the naïve CD4+ T cells isolation kit II (Miltenyi Biotec, Auburn, CA) and plated on anti-CD3- (3 μg ml−1) coated plates with and without TL1A (300 ng ml−1) for 48 h. Medium was harvested for Luminex 27 Bioplex analysis (Bio-Rad, Hercules, CA) and/or IL2 AlphaLISA and the cells were harvested for Taqman analysis. For CD161+ studies, total PBMCs were isolated from HVs and treated with IL12, IL18, and TL1A (200 ng ml−1) or TNFα (200 ng /ml−1; Peprotech, Rocky Hill, NJ) for 18 h, followed by the addition of GolgiPlug (BD Biosciences) for 4 h. Following study approval by Independent IRB (Sunrise, CA), whole blood was collected from CD patient with active disease by Bioreclamation, LLC (Liverpool, NY), following patient consent. Samples were shipped for same-day delivery and processed simultaneously with HV’s peripheral blood. For flow cytometry, cells were stained with surface markers, including CD3, CD4, CD56, CD161, CD45RA, DR3, DR3Fc, DcR3-Fc, CCR6, TNFRI, and TNFRII, and intracellular IFNγ and TNFα were stained using Cytofix/Cytoperm Plus kit (BD Biosciences, San Jose, CA). Anti-human TNFα (R&D System) and anti-mouse TNFα (Leinco, St Louis, MO) were used to neutralize TNFα levels. To isolate CD161+ cells, T cells were labeled with anti-CD161 fluorescein isothiocyanate and then CD161+ and CD161− cells were sorted using Aria sorter (BD Biosciences). Medium was harvested for Luminex 27 Bioplex analysis (Bio-Rad). For all in vitro human assays, each experiment was performed with a minimum of three different human donors and with an n=2–8 replicates per sample. P-values were generated with Student’s t-test.
Immunofluorescent labeling of TL1A on activated human monocytes. Human monocytes isolated from PBMC were plated in 12-well plates with plate-bound, crosslinked human IgG for overnight. Cells were then stained with mouse anti-human TL1A monoclonal antibody-0011 at a final concentration of 1 μg ml−1. Secondary detection was facilitated using a 1/200 dilution of AlexaFluor 488-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Slides were examined using a Zeiss Axioplan II microscope and digital images captured using Zeiss Axiovision software (Oberkochen, Germany).
Generation of human soluble and membrane TL1A. For the generation of recombinant human soluble TL1A, amino acids 72–251 were cloned with the N-terminal tag composed of AviTag, six histidine residues, and a thrombin-cleavage site under the control of the cytomegalovirus promoter. The resulting plasmid was transiently transfected into FreeStyle 293-F cells (Invitrogen, Grand Island, NY). Cell culture supernatants were harvested 7 days after transfection and purified by immobilized metal ion affinity chromatography. For the expression of MbTL1A, the construct was generated by replacing amino acids 67–94 (VQFQALKGQEFAPSHQQVYAPLRADGDK) within human TL1a by a flexible (G4S)4 linker (GGGGSGGGGSGGGGSGGG GS). CHO cells (ATCC, Manassas, VA) were transfected, and after 3 weeks of transfection, single cells were sorted and checked for TL1a expression by FACS analysis.
Generation of mouse anti-human TL1A antibody. NMRI mice received six consecutive immunizations of full-length TL1A DNA at days 0, 14, 28, 42, 56, and 70. Animals with highest titers were selected for boosting at day 96 by intravenous injection of 25 μg of recombinant human soluble TL1a (R&D System) and monoclonal antibodies were isolated by hybridoma technology.
Patient populations. Patients with a diagnosis of CD, UC, and non-IBD controls were included in the study after obtaining written informed consent. The study was approved by the Institutional Ethics Committee of the Hospital Clinic (Barcelona, Spain) and was performed in accordance with the principles stated in the Declaration of Helsinki. Non-IBD controls were those subjects undergoing colonoscopy for mild gastrointestinal symptoms or a screening for colorectal cancer, and who presented no lesions during examination. Inclusion criteria for CD and UC patients were: age between 18 and 65 years, and mucosal lesions at the time of study. Endoscopic disease activity was assessed using the CD endoscopic activity index and the endoscopic Mayo subscore for CD and UC, respectively. Patients with concomitant infections were excluded. Serum was obtained from peripheral blood (5 ml) drawn from a total of 42 CD, 17 UC, and 21 non-IBD controls. Clinical and demographic characteristics of the subjects are shown in Table 1.
Microarray data analysis. Microarray raw data was analyzed using Bioconductor tools in R (www.r-project.org). The robust multiarray average algorithm was applied to Affymetrix raw data files to normalize them, resulting in a log 2 expression value for each probe set. Differential expression analysis was assessed by using linear models for microarray data, based on empirical Bayes moderated t-statistics for selected probes. To correct for multiple testing, the false discovery rate was estimated from P-values derived from the moderated t-statistics using the method of Benjamini and Hochberg. Genes were considered significantly up- or downregulated if they had a P-value <0.05 and at least a 1.5-fold variation in mean expression (log 2 FC>|0.5849|).
qPCR of CD patient biopsies. This study was approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals and patient recruitment was between October 2008 and October 2012. Informed consent was obtained from all research participants. A single gastroenterologist was responsible for assessment of the IBD patients (FS). Biopsies were taken from a predefined and consistent location immediately proximal to the junction of the sigmoid colon and rectum. For RNA analysis, a single biopsy was immediately placed and stored overnight in RNALater (Ambion, Poole, Dorset, UK) at 4 °C. Following overnight saturation, the biopsies were placed in fresh RNALater and stored at −80 °C. Total RNA was isolated from sigmoidal biopsies using the mirVana miRNA Isolation Kit (Ambion, Foster City, CA). cDNA was synthesized using 1 μg total RNA. All PCR reactions were performed in duplicate using TaqMan assays (Applied Biosystems, Life Technologies, Foster City, CA) in 384-well plates on the LightCycler 480 system (Roche Applied Science, Indianapolis, IN). Statistical significance was determined with one-way analysis of variance with post hoc analysis. All statistical tests were performed using commercially available statistic software (GraphPad Software, La Jolla, CA). Data are represented by mean±s.e.m., unless otherwise stated. P-values of <0.05 were considered significant (*P<0.05, **P<0.01, and ***P<0.001).
References
Migone, T.S. et al. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity 16, 479–492 (2002).
Meylan, F., Richard, A.C. & Siegel, R.M. TL1A and DR3, a TNF family ligand-receptor pair that promotes lymphocyte costimulation, mucosal hyperplasia, and autoimmune inflammation. Immunol Rev 244, 188–196 (2011).
Meylan, F. et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity 29, 79–89 (2008).
Prehn, J.L., Thomas, L.S., Landers, C.J., Yu, Q.T., Michelsen, K.S. & Targan, S.R. The T cell costimulator TL1A is induced by FcgammaR signaling in human monocytes and dendritic cells. J Immunol 178, 4033–4038 (2007).
Shih, D.Q. et al. Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. Eur J Immunol 39, 3239–3250 (2009).
Shih, D.Q. et al. Insights into TL1A and IBD pathogenesis. Adv. Exp. Med. Biol. 691, 279–288 (2011).
Shih, D.Q., Targan, S.R. & McGovern, D. Recent advances in IBD pathogenesis: genetics and immunobiology. Curr Gastroenterol Rep 10, 568–575 (2008).
Michelsen, K.S. et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One 4, e4719 (2009).
Yamazaki, K. et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet 14, 3499–3506 (2005).
Kakuta, Y., Kinouchi, Y., Negoro, K., Takahashi, S. & Shimosegawa, T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut 55, 1527–1528 (2006).
Kakuta, Y. et al. TNFSF15 transcripts from risk haplotype for Crohn’s disease are overexpressed in stimulated T cells. Hum Mol Genet 18, 1089–1098 (2009).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40, 955–962 (2008).
Bamias, G. et al. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol 171, 4868–4874 (2003).
Prehn, J.L. et al. Potential role for TL1A, the new TNF-family member and potent costimulator of IFN-gamma, in mucosal inflammation. Clin Immunol (Orlando, FL) 112, 66–77 (2004).
Bamias, G. et al. Differential expression of the TL1A/DcR3 system of TNF/TNFR-like proteins in large vs. small intestinal Crohn’s disease. Dig Liver Dis 44, 30–36 (2012).
Bamias, G., Siakavellas, S.I., Stamatelopoulos, K.S., Chryssochoou, E., Papamichael, C. & Sfikakis, P.P. Circulating levels of TNF-like cytokine 1A (TL1A) and its decoy receptor 3 (DcR3) in rheumatoid arthritis. Clin Immunol (Orlando, FL) 129, 249–255 (2008).
Bamias, G. et al. High intestinal and systemic levels of decoy receptor 3 (DcR3) and its ligand TL1A in active ulcerative colitis. Clin Immunol (Orlando, FL) 137, 242–249 (2010).
Murphy, S.F., Kwon, J.H. & Boone, D.L. Novel players in inflammatory bowel disease pathogenesis. Curr Gastroenterol Rep 14, 146–152 (2012).
Pineton de Chambrun, G.P. & Sandborn, W.J. IBD in 2011: advances in IBD management—towards a tailored approach. Nat Rev Gastroenterol Hepatol 9, 70–72 (2012).
Melmed, G.Y. & Targan, S.R. Future biologic targets for IBD: potentials and pitfalls. Nat Rev Gastroenterol Hepatol 7, 110–117 (2010).
Biener-Ramanujan, E., Gonsky, R., Ko, B. & Targan, S.R. Functional signaling of membrane-bound TL1A induces IFN-gamma expression. FEBS Lett 584, 2376–2380 (2010).
Papadakis, K.A. et al. TL1A synergizes with IL-12 and IL-18 to enhance IFN-gamma production in human T cells and NK cells. J Immunol 172, 7002–7007 (2004).
Papadakis, K.A. et al. Dominant role for TL1A/DR3 pathway in IL-12 plus IL-18-induced IFN-gamma production by peripheral blood and mucosal CCR9+ T lymphocytes. J Immunol 174, 4985–4990 (2005).
Monteleone, G. et al. Interleukin 12 is expressed and actively released by Crohn’s disease intestinal lamina propria mononuclear cells. Gastroenterology 112, 1169–1178 (1997).
Maerten, P. et al. Involvement of interleukin 18 in Crohn’s disease: evidence from in vitro analysis of human gut inflammatory cells and from experimental colitis models. Clin Exp Immunol 135, 310–317 (2004).
Okazawa, A. et al. Th1-mediated intestinal inflammation in Crohn’s disease may be induced by activation of lamina propria lymphocytes through synergistic stimulation of interleukin-12 and interleukin-18 without T cell receptor engagement. Am J Gastroenterol 97, 3108–3117 (2002).
Siegmund, B. et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-gamma and TNF-alpha production. Am J Physiol Regul Integr Comp Physiol 281, R1264–R1273 (2001).
O’Keeffe, J. et al. Diverse populations of T cells with NK cell receptors accumulate in the human intestine in health and in colorectal cancer. Eur J Immunol 34, 2110–2119 (2004).
Takahashi, T., Dejbakhsh-Jones, S. & Strober, S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol 176, 211–216 (2006).
Kleinschek, M.A. et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med 206, 525–534 (2009).
Maggi, L. et al. Distinctive features of classic and non-classic (Th17-derived) human Th1 cells. Eur J Immunol e-pub ahead of print (2012).
Wen, L., Zhuang, L., Luo, X. & Wei, P. TL1A-induced NF-kappaB activation and c-IAP2 production prevent DR3-mediated apoptosis in TF-1 cells. J Biol Chem 278, 39251–39258 (2003).
Ji, C. et al. CD4-anchoring HIV-1 fusion inhibitor with enhanced potency and in vivo stability. J Biol Chem 284, 5175–5185 (2009).
Meylan, F. et al. The TNF-family cytokine TL1A drives IL-13-dependent small intestinal inflammation. Mucosal Immunol 4, 172–185 (2011).
Shih, D.Q. & Targan, S.R. Insights into IBD pathogenesis. Curr Gastroenterol Rep 11, 473–480 (2009).
Acknowledgements
We acknowledge Dr Al-Shamkhani for the mouse anti-TL1A antibody. We thank MD Biosciences for running the TNBS model and subsequent analysis. We thank Gang Chen for project support. Finally, we thank Alain Tissot for project insight and support and Chris Kitson, Franck Barrat, and Thomas Tan for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Jin, S., Chin, J., Seeber, S. et al. TL1A/TNFSF15 directly induces proinflammatory cytokines, including TNFα, from CD3+CD161+ T cells to exacerbate gut inflammation. Mucosal Immunol 6, 886–899 (2013). https://doi.org/10.1038/mi.2012.124
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2012.124
This article is cited by
-
TNFSF15 promoter polymorphisms increase the susceptibility to small cell lung cancer: a case-control study
BMC Medical Genetics (2019)
-
Oestrogen-deficiency induces bone loss by modulating CD14+ monocyte and CD4+ T cell DR3 expression and serum TL1A levels
BMC Musculoskeletal Disorders (2019)
-
At the Junction of Immunity and Barrier Function: The Immunomodulatory Protein TL1A May Also Regulate Intestinal Permeability
Digestive Diseases and Sciences (2019)
-
Comparative Network Analysis of Patients with Non-Small Cell Lung Cancer and Smokers for Representing Potential Therapeutic Targets
Scientific Reports (2017)
-
TNF superfamily cytokines in the promotion of Th9 differentiation and immunopathology
Seminars in Immunopathology (2017)