Abstract
Gingival epithelia utilize multiple signaling pathways to regulate innate immune responses to various oral bacteria, but little is understood about how these bacteria alter epithelial epigenetic status. In this study we report that DNA methyltransferase (DNMT1) and histone deacetylase expression were decreased in gingival epithelial cells treated with oral pathogen Porphyromonas gingivalis and nonpathogen Fusobacterium nucleatum. Pretreatment with trichostatin A and sodium butyrate, which increase acetylation of chromatin histones, significantly enhanced the gene expression of antimicrobial proteins human β-defensin 2 (hBD2) and CC chemokine ligand 20 (CCL20) in response to both bacterial challenges. Pretreatment with DNMT inhibitor 5′-azacytidine increased hBD2 and CCL20 expression in response to F. nucleatum, but not to P. gingivalis. Furthermore, we observed a differential pattern of protein levels of H3K4me3, which has been associated with chromatin remodeling and activation of gene transcription, in response to P. gingivalis vs. F. nucleatum. This study provides a new insight into the bacteria-specific innate immune responses via epigenetic regulation.
Similar content being viewed by others
Introduction
Epigenetics is heritable and reversible changes in gene expression without altering DNA sequence. The mechanisms of epigenetic modifications include histone acetylation, histone methylation, and DNA methylation, and these modifications provide a way to control the expression of genes involved in various cellular functions.1, 2 Enzymes involved in these epigenetic mechanisms are: histone acetyltransferases, histone deacetylases (HDACs), histone methyltransferases, and DNA methyltransferases (DNMTs). When histones are acetylated, transcription factors can access DNA, leading to gene transcription, whereas deacetylated histones lead to condensed (or closed) chromatin structure, making DNA inaccessible to transcription factors and preventing gene expression. In addition, histone methylation has a pivotal role in the maintenance of both active and suppressed states of gene expression, depending on the sites of methylation. The methylation of histone H3 at lysine-4 and lysine-36 (H3K4 and H3K36) is implicated in activation of transcription, whereas methylation of histone H3 at lysine-9 and lysine-27 (H3K9 and H3K27) is correlated with repression of transcription.3 Perturbed balance between acetylation/deacetylation or methylation/demethylation leads to aberrant expression of key regulators of cell development, disease, and more often cancer development. Most of epigenetic studies thus far have focused on cancer progression, but the role of host inflammation on modification of epigenetic patterns is still in its infancy. Periodontitis results in an inflammatory reaction of the periodontium to pathogenic microorganisms. A number of Gram-negative anaerobic bacteria, including Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, are associated with periodontal disease.4 In addition to bacterial infection, genetic and/or environmental factors contribute to occurrence and progression of periodontal disease.5, 6 Many patients with the same clinical symptoms respond differently to the same therapy, suggesting interindividual variability observed as the clinical outcome of the disease is influenced by genetic7, 8 as well as epigenetic factors.9 Recent studies suggest that epigenetics have a critical role in regulating inflammatory responses and that the manifestation and severity of periodontal disease may be influenced by epigenetic factors.9, 10
The innate immune system is the first line of defense against invading microorganism by producing antimicrobial peptides, such as human β-defensins (hBDs), which are widely expressed in epithelial tissue including oral cavity. They have a broad spectrum of activity against Gram-negative and Gram-positive bacteria as well as some fungi and virus.11 In addition to their direct effect, hBDs also stimulate antigen-presenting dendritic cells and link other innate immune components or adaptive immune responses, resulting in efficient and coordinated epithelial barrier function against invading bacteria.12 Therefore, it seems reasonable that periodontal disease could be prevented or mitigated by induction of host innate immune function.
We have previously reported that exposure of gingival epithelial cells (GECs) to different oral bacteria leads to differential induction of downstream innate immune markers and that the signaling pathways utilized also differ between bacteria.13, 14 Therefore, we hypothesize that differential epigenetic changes will result from the presence of different oral bacteria and that these epigenetic changes could influence the innate immune responses in the host. The goal of this study is to investigate how epigenetic modifications brought on by exposure to oral bacteria, including a periodontal pathogen, affect host innate immune responses, such as hBD2 and CC chemokine ligand 20 (CCL20) expression.
Results
Changes in HDAC1, HDAC2, and DNMT1 in response to the presence of P. gingivalis and F. nucleatum
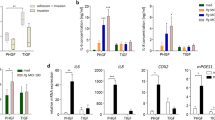
We first investigated any changes in the expression levels of chromatin-remodeling enzymes after GECs were exposed to oral bacteria. HDACs remove acetyl groups from histone, leading to suppression of genes. DNMTs catalyze transfer of methyl groups onto DNA, which also leads to gene suppression. First, the expression levels of HDAC1, HDAC2, and DNMT1 were investigated in human immortalized keratinocyte cell line (TERT) at different time points (4 and 24 h) and multiplicities of infection (MOIs; 10:1 and 100:1). Quantitative real-time PCR (QRT-PCR) analyses showed changes in the expression levels of DNMT1, HDAC1, and HDAC2 when TERT cells were treated with P. gingivalis (a pathogen) or F. nucleatum (a bridging organism between pathogens and nonpathogens) at MOIs of 10:1 and 100:1 (Figure 1) for 4 and 24 h. The gene expression of HDAC1 was decreased significantly at MOI 100:1 for 24 h in cells treated with P. gingivalis, but not with F. nucleatum. The gene expression of HDAC2 slightly increased with F. nucleatum at 4 h for MOI 10:1. Significant decreases of DNMT1 gene expression levels were observed in TERT cells treated with both bacteria at MOI of 100:1 for 24 h. As more significant changes were observed at 24 h, we further compared these results from human immortalized cell line with human primary GECs at 24 h and various MOIs (Figure 2). The expression level of DNMT1 decreased in response to both P. gingivalis and F. nucleatum (P=0.003 and 0.006, respectively; Figure 2a). This decrease was observed at all MOIs tested in response to both P. gingivalis and F. nucleatum. This decrease in DNMT1 gene expression was dose dependent in GECs in response to F. nucleatum. The gene expression level of HDAC1 significantly decreased in GECs when the cells were treated with P. gingivalis for 24 h at all MOIs compared with unstimulated control (P=0.001; Figure 2b). On the other hand, the expression of HDAC1 significantly decreased in GECs to the lowest level when treated with F. nucleatum for 24 h at MOI 10:1 (P=0.03; Figure 2b). The gene expression of HDCA2 significantly decreased in GECs in response to F. nucleatum at MOIs 50:1 and 100:1, whereas only at MOI 100:1 in response to P. gingivalis (Figure 2c). The decrease of DNMT1 and HDAC2 gene expression showed similar trends in GECs compared with what we observed in TERT cells, whereas the expression of HDAC1 in response to P. gingivalis and F. nucleatum differed between the two cell types. These data indicate that the gene expression of these chromatin-remodeling enzymes may have cell type-specific responses.
Differential mRNA expression of HDAC1, HDAC2 and DNMT1 in human TERT cells in response to oral bacteria. Differential mRNA expression of (a) histone deacetylases 1 and 2 (HDAC1 and HDAC2) and (b) DNA methyltransferase (DNMT1) in human TERT cells in response to Porphyromonas gingivalis vs. Fusobacterium nucleatum. Human TERT cells were stimulated with P. gingivalis (Pg) or F. nucleatum (Fn) at multiplicities of infection (MOIs) of 10:1, 50:1, and 100:1 for 4 or 24 h. Unstimulated cells at 4 and 24 h served as controls. Changes in mRNA expression were evaluated by quantitative real-time PCR (QRT-PCR) and results are expressed as fold change in gene expression compared with the corresponding unstimulated controls (4 and 24 h) after normalization with glyceraldehydes-3-phosphate dehydrogenase (GAPDH). The experiment was repeated twice using TERT cells. Error bars indicate s.e.m. Asterisks indicate statistically significant difference compared with unstimulated control (Ctl) (*P<0.05, **P<0.01).
Differential decreased mRNA expression of HDAC1, HDAC2 and DNMT1 in gingival epithelial cells in response to various doses of oral bacteria. mRNA expression of (a) DNA methyltransferase (DNMT1), (b) histone deacetylase 1 (HDAC1), and (c) histone deacetylase 2 (HDAC2) are differentially decreased in gingival epithelial cells (GECs) in response to various doses of Porphyromonas gingivalis vs. Fusobacterium nucleatum. GECs were stimulated with P. gingivalis (Pg) or F. nucleatum (Fn) at multiplicities of infection (MOIs) of 10:1, 50:1, 100:1, and 200:1 for 24 h. Changes in mRNA expression were evaluated by quantitative real-time PCR (QRT-PCR) and results are expressed as fold change in gene expression compared with the unstimulated control after normalization with the housekeeping gene glyceraldehydes-3-phosphate dehydrogenase (GAPDH). The data are derived from three different cell donors tested in duplicate. Error bars indicate s.e.m. Asterisks indicate statistically significant difference compared with unstimulated control (Ctl) (*P<0.05, **P<0.01).
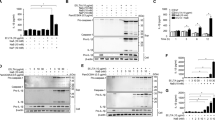
We further confirmed the gene expression of chromatin-remodeling enzymes (HDAC1, HDAC2, and DNMT1) in GECs treated with oral bacteria at the protein levels using western blot analyses. P. gingivalis (MOI 100:1) significantly decreased HDAC1, HDAC2, and DNMT1 proteins in GECs compared with the unstimulated controls at 24 h. In contrast, F. nucleatum (MOI 100:1) did not show any effects on the expression of these proteins in GECs at 24 h compared with controls (Figure 3). The analysis of the protein expression pattern of DNMT1, HDAC1, and HDAC2 followed the same trend as the mRNA expression in GECs treated with P. gingivalis.
Protein levels of histone deacetylases 1 and 2 (HDAC1 and HDAC2) and DNA methyltransferase (DNMT1) are differentially expressed in gingival epithelial cells (GECs) in response to Porphyromonas gingivalis (Pg) and Fusobacterium nucleatum (Fn). GECs were stimulated with P. gingivalis (Pg) or F. nucleatum (Fn) at multiplicities of infection (MOIs) of 100:1 for 24 h. Nuclear proteins were extracted, denatured at 70 °C for 10 min, and separated by NuPAGE electrophoresis system. Nuclear extracts of Hela cells probed with individual primary antibody were used as positive controls. The data are derived from two different cell donors tested in duplicate.
Gene expression of epithelial innate immune markers is increased following the inhibition of HDAC and DNMT
β-Defensins and CCL20 have been known as critical markers of innate immune responses against microbial invasion.15, 16 We previously reported that P. gingivalis and F. nucleatum are all excellent inducers of hBD-213 and CCL20 (also known as MIP3α (macrophage inflammatory protein-3α)).17 In order to study the downstream response of innate immune markers following epigenetic modifications, we tested the effect of the inhibitors for chromatin-modifying enzymes followed by bacterial stimulation on the expression of the antimicrobial proteins hBD-2 and CCL20. GECs were pretreated with HDAC inhibitors sodium butyrate (SB) or trichostatin A (TSA), or DNMT inhibitor 5′-azacytidine (AZA) before bacterial exposure. Treatment of GECs with AZA demethylates DNA, leading to gene activation, whereas exposure to SB or TSA results in a higher level of histone acetylation. The gene expression of hBD2 and CCL20 was significantly increased in GECs in response to both P. gingivalis and F. nucleatum, which is consistent with our previous report (Figures 4 and 5).13, 18 Treatment with inhibitors alone did not significantly change expression of hBD2 and CCL20 in GECs. The inhibitor SB had no effect on the gene expression of hBD2 and CCL20 in response to these oral bacteria compared with relative bacterial treatment alone. Treatment with TSA, another HDAC inhibitor structurally unrelated to SB, resulted in significantly increased hBD2 and CCL20 gene expression in GECs in response to both oral bacteria (Figures 4b and 5b) compared with the bacterial treatment alone. Combining inhibitors SB and TSA synergistically enhanced CCL20 expression in GECs (Figure 5c). Treatment with HDAC inhibitors amplified hBD2 and CCL20 gene expression, thus suggesting that increasing histone acetylation will aid innate immune responses in GECs in the presence of oral bacteria. Treatment with AZA significantly increased gene expression of hBD2 and CCL20 in GECS in response to F. nucleatum, but not to P. gingivalis (Figures 4d and 5d). Our data suggest that the oral bridging microorganism F. nucleatum induced hBD2 and CCL20 via both demethylation and acetylation mechanisms, whereas the induction by oral pathogen P. gingivalis is only via acetylation mechanism. Thus, taken together, our data suggest that epithelial innate immune responses (hBD2 and CCL20) are regulated by epigenetic modifications, and these responses are bacteria specific.
mRNA expression of innate immune markers human β-defensin 2 (hBD-2) are increased when histone deacetylase (HDAC) and DNA methyltransferase (DNMT) are inhibited. Gingival epithelial cells (GECs) were pretreated with trichostatin A (TSA; 9 and 45 mm), sodium butyrate (SB; 0.5 and 2.0 mm) or 5′-azacytidine (AZA; 1 and 10 μm) for 4 h, and subsequently exposed to Porphyromonas gingivalis (multiplicity of infection (MOI) 100:1) or Fusobacterium nucleatum (MOI 100:1) for 16 h. Gene expression of hBD2 was evaluated by quantitative real-time PCR (QRT-PCR) compared with unstimulated control after normalization with glyceraldehydes-3-phosphate dehydrogenase (GAPDH). Controls include unstimulated control, bacteria-alone treatment, and various inhibitors alone as indicated. No significant changes were found in the gene expression of hBD2 in GECs treated with inhibitor only: (a) SB, (b) TSA, (c) SB+TSA, and (d) AZA compared with unstimulated control. Data are expressed as means of fold change±s.e.m. from three donors evaluated in duplicate. Asterisks indicate statistically significant difference compared with unstimulated control (Ctl) (*P<0.05, **P<0.01).
mRNA expression of innate immune markers CC chemokine ligand 20 (CCL20) are increased when histone deacetylase (HDAC) and DNA methyltransferase (DNMT) are inhibited. Gingival epithelial cells (GECs) were pretreated with trichostatin A (TSA; 9 and 45 mm), sodium butyrate (SB; 0.5 and 2.0 mm), or 5′-azacytidine (AZA; 1 and 10 μm) for 4 h, and subsequently exposed to Porphyromonas gingivalis (multiplicity of infection (MOI) 100:1) or Fusobacterium nucleatum (MOI 100:1) for 16 h. Gene expression of (b) CCL20 was evaluated by quantitative real-time PCR (QRT-PCR) compared with unstimulated control after normalization with glyceraldehydes-3-phosphate dehydrogenase (GAPDH). Controls include unstimulated control, bacteria-alone treatment, and various inhibitors alone as indicated. No significant changes were found in the gene expression of CCL20 in GECs treated with inhibitor only: (a) SB, (b) TSA, (c) SB+TSA, and (d) AZA compared with unstimulated control. Data are expressed as means of fold change±s.e.m. from three donors evaluated in duplicate. Asterisks indicate statistically significant difference compared with unstimulated control (Ctl) (*P<0.05, **P<0.01).
Differential increase of interleukin-8 (IL-8) secretion following the inhibition of HDAC and DNMT
To determine whether an increase in histone acetylation or demethylation would also induce additional immune responses in GECs, IL-8 secretion was measured (Figure 6). Stimulation with F. nucleatum resulted in the increased level of IL-8 secretion, whereas P. gingivalis decreased IL-8 secretion, which is in agreement with our earlier finding. IL-8 secretion was significantly increased when GECs were pretreated with HDAC inhibitor SB or TSA followed by exposure to P. gingivalis, but not to F. nucleatum, compared with the bacteria treatment only (Figure 6a, b). IL-8 secretion significantly increased when GECs were pretreated with the combination of low doses of SB and TSA in response to P. gingivalis and F. nucleatum, which was similar to the observation in CCL20 expression (Figure 5c). When GECs were pretreated with DNMT1 inhibitor AZA, no changes in IL-8 secretion were observed in the presence of two bacteria (Figure 6d). These data suggest that IL-8 secretion may be related to acetylation status in GECs.
Interleukin-8 (IL-8) secretion is increased when histone deacetylase (HDAC) and DNA methyltransferase (DNMT) are inhibited. Gingival epithelial cells (GECs) were pretreated with trichostatin A (TSA; 9 and 45 mm), sodium butyrate (SB; 0.5 and 2.0 mm), or 5′-azacytidine (AZA; 1 and 10 μm) for 4 h, and subsequently exposed to Porphyromonas gingivalis (multiplicity of infection (MOI) 100:1) or Fusobacterium nucleatum (MOI 100:1) for 16 h. Secretion of IL-8 in response to various oral bacteria is evaluated by enzyme-linked immunosorbent assay (ELISA). Cell-free supernatant was collected and the amount of IL-8 secreted is shown in pg ml–1. Unstimulated cells (UN) are used as controls in each experiment. Data from duplicates with cells from three different donors are shown. No significant changes were found in the secretion of IL-8 in GECs treated with inhibitor only: (a) SB, (b) TSA, (c) SB+TSA, and (d) AZA compared with unstimulated control. Asterisks indicate statistically significant difference compared with unstimulated control (Ctl) (*P<0.05, **P<0.01).
Tri-methyl-histone H3 protein level is differentially regulated by P. gingivalis and F. nucleatum in GECs
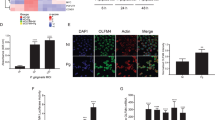
Histone methylation has a pivotal role in the maintenance of both active and suppressed states of gene expression, depending on the sites of methylation. Methylation of Lys4 of histone 3 is generally associated with transcriptionally active chromatin.19 In order to evaluate the changes in histone H3 methylation level in GECs when exposed to bacteria, we detected endogenous levels of histone H3 tri-methylated at Lys4 (H3K4me3) following exposure to P. gingivalis or F. nucleatum for 24 h at MOI 100:1. The protein amount was significantly decreased when GECs were stimulated with P. gingivalis compared with unstimulated control, whereas the level of this protein had no significant change after exposure to F. nucleatum (Figure 7) compared with the control. These results demonstrate differential induction of H3K4 methylation and further suggest that oral bacterial infection is associated with changes in H3K4 methylation pattern.
Protein levels of histone H3 methylated at Lys4 were evaluated with PathScan enzyme-linked immunosorbent assay (ELISA). Gingival epithelial cells (GECs) were exposed to Porphyromonas gingivalis or Fusobacterium nucleatum for 24 h at multiplicity of infection (MOI) of 100:1, and then nuclear protein was extracted followed by sonication. The H3 tri-methylated at Lys4 was captured by coated antibody after incubation with cell lysates, and histone H3 protein level was quantified according to the absorbance readings at 450 nm. Protein expression was expressed as the ratio of absorbance readings normalized to relative protein amount. The data are average from three different donor cells tested with standard error deviation. The asterisks indicate the significant difference vs. the respective unstimulated control (**P<0.01).
Epigenetic modification of genes, whose function is associated with growth control and inflammation, is differentially regulated by P. gingivalis and F. nucleatum
Gene promoter methylation is one of the most common epigenetic mechanisms to silence gene expression. In order to identify specific methylated genes associated with P. gingivalis or F. nucleatum infection, we examined the changes in methylation levels of 24 different cytokine-producing genes in TERT cells after the cells were exposed to P. gingivalis or F. nucleatum for 24 h at MOIs 10:1, 100:1, and 200:1 (Table 1). A dose-dependent increase in methylation levels of the following genes was observed when GECs were exposed to P. gingivalis: CD276, an immune regulator (P<0.005); elastase 2, a serine protease that has a role in inflammatory diseases (P<0.01); INHBA (inhibin, β), a tumor-suppressing protein (P<0.01); GATA3 (GATA binding protein 3), a putative tumor suppressor (P<0.005); Toll-like receptor 2 (TLR2; P<0.005); and IL-12A (P<0.005). Stimulation of GECs with P. gingivalis also resulted in hypomethylation of ZNF287, a member of zinc-finger protein family, and signal transducer and activator of transcription 5A (STAT5A; P<0.005), which mediates cellular responses to cytokines IL-2, IL-3, IL-7, and granulocyte–macrophage colony-stimulating factor. On the other hand, the methylation levels of elastase 2 (P<0.005) and GATA3 (P<0.01) decreased after cells were stimulated with F. nucleatum. Interestingly, only F. nucleatum induced hypermethylation of MALT1 (mucosa-associated lymphoid tissue lymphoma translocation gene) in GECs.
We have observed that hBD2, CCL20, and chemokine IL-8 are differentially modulated via epigenetic mechanism, and also found that multiple genes are differentially methylated after exposure to P. gingivalis or F. nucleatum. In order to further understand biological correlation among the genes that respond to the bacterial treatments, we used PubGene to identify biological relationship based on literature co-occurrence index (Figure 8). The queried genes DEFB4 (hBD2), CCL20, and IL-8 are strongly associated with the genes that showed hypermethylation (ELA2, TRL2, STAT5A, IL-12A, and GATA3) based on literature co-citation. This illustrates the complex coregulatory patterns of tested genes, and highlights the biological relevance for the tested genes.
Relevant gene networks based on literature co-occurrence. We submitted queried genes DEFB4 (human β-defensin 2 (hBD2)), CC chemokine ligand 20 (CCL20), interleukin-8 (IL-8), and methylated genes in Table 1 to the PubGene and identified coexpressed possibility based on the literature co-occurrence. Red color represents query genes, purple color represents the first-order literature, and black color represents the second-order literature. The color reproduction of this figure is available on the html full text version of the manuscript.
Genes associated with chromatin modification changed in host cells that were treated with different oral bacteria
In order to broaden our understanding of which genes are involved with chromatin modifications in epithelial responses to bacteria, we performed microarray ontology analysis (BRB Array tool)20 based on our previous study and on public data sets from NCBI (National Center for Biotechnology Information) Gene Expression Omnibus (accession number GSE6927).21, 22, 23 A number of genes that are associated with chromatin modification were altered in host cells that were treated with P. gingivalis or F. nucleatum (Table 2). The changes observed in histone deacetylases and DNA methyltransferases in response to P. gingivalis correspond to our mRNA data (Figures 1 and 3). Thus, our findings using different molecular methods are consistent and suggest that various genes associated with epigenetic modifications are differentially regulated when oral epithelial cells are exposed to oral bacteria.
Discussion
In this study, we showed that the presence of bacteria results in epigenetic modifications in gingival epithelia. We also demonstrated that epigenetic modifications have a critical role in the regulation of innate immune responses of gingival epithelia. Epigenetic modifications, such as DNA methylation and histone acetylation or methylation status, lead to altered gene expression and have been associated with cancer development and tumor growth. However, the role of epigenetic mechanism in host innate immune responses to bacteria–host interactions has not been studied. Thus, our study addresses a novel and an important question of the role of epigenetics in epithelial innate immunity. Our data suggest that exposure to different oral bacteria results in differential methylation profile, and bacteria-induced expression of epithelial antimicrobial molecules hBD2 and CCL20 is regulated through epigenetic mechanisms in GECs.
Factors that influence epigenetic profile are wide ranging and include environmental toxins, individual diet, as well as microbial infections. Gastric cancer is one type of disease that has been identified as resulting from epigenetic modification following bacterial infection.24 In the oral cavity, epithelial tissues are constantly exposed to a variety of bacteria, and different bacteria elicit different innate immune responses from the host. These divergences may be seen between pathogens vs. nonpathogens and/or between various species of pathogens. We have previously reported that pathogens and nonpathogens utilize different signaling pathways in the induction of epithelial innate immune markers.13, 14 Because of the high number of bacteria present in the oral cavity at all times, we strongly suspected that the presence of bacteria will influence the epigenetic status of the host. Our data showed that stimulation of GECs with P. gingivalis resulted in hypomethylation of ZNF287. The upregulation of ZNF287 has been associated with cardiovascular disease;25 thus, our observation is in line with recent studies linking periodontal disease with increased risk of systemic disease.26 On the other hand, F. nucleatum, but not P. gingivalis, induced hypermethylation of MALT1 that stimulates IKK catalytic activity, resulting in nuclear factor (NF-κB) activation.27 The methylation of MALT1 is associated with the silencing of MALT1 gene, which is consistent with our previous data that F. nucleatum does not utilize the NF-κB signaling pathway in the induction of innate immune responses.13, 14 Taken together, our data suggest that oral epithelia respond to the presence of different bacteria via differential epigenetic changes on various genes.
We showed both species of oral bacteria tested significantly decreased gene expression of chromatin-modifying enzyme DNMT1 in GECs. In contrast, we also observed that the two oral bacteria differentially induced hypermethylation of specific genes in GECs as listed in Table 1. The explanation for this discrepancy is that DNMT1 has an ability to promote only 20% of overall genomic methylation, although it is a major DNMT in humans.28 In addition, two other enzymes, DNMT3a and DNMT3b, have also been shown to possess DNMT activity;29 thus, other methyltransferases may respond the same way.
HDAC inhibitors are known as agents that modulate the expression of genes by increasing histone acetylation, thereby regulating chromatin structure, transcription, and increasing antimicrobial function of cathelicidin expression in skin keratinocytes. The similar phenomenon was also observed in gingival epithelium. In this study, we have found that inhibition of HDAC (SB) has no significant effects on GECs in response to both oral bacteria tested in terms of gene expression of hBD2 and CCL20, whereas inhibition by TSA significantly induced gene expression in GECs in response to these bacteria. This may be because of the fact that TSA is a more potent HDAC inhibitor. More importantly, inhibition by both SB and TSA synergistically stimulated the gene expression of CCL20 in GECs in response to oral bacteria, further confirming that innate immune responses to oral bacteria are dependent on the deacetylation status of host.
HBD2 and CCL20 with innate immune properties have been well studied. HBD2, an inducible antimicrobial peptide, has been found to be active against a panel of oral bacteria and Candida species.11, 15 CCL20, a chemokine with antimicrobial regions structurally related to hBD2, has antimicrobial activity mainly against Gram-negative bacteria and has a critical role in pulmonary innate immunity.30 Therefore, the present finding suggested that altered epigenetic modification could potentially increase antimicrobial activity and enhance innate immunity in GECs.
Recent studies have demonstrated that methylation of histone H3 at K4 is important for the initial activation of transcription, because methylation of histone H3 at K4 precedes histone acetylation.19 Our data showed that P. gingivalis significantly decreased tri-methylation histone H3 at K4 protein expression, but F. nucleatum did not, indicating that P. gingivalis could suppress the activation of transcription, consistent with the “stealth-like” properties of P. gingivalis.31 Our previous finding also support that the secretion level of cytokines and chemokines is lower in P. gingivalis-stimulated dendritic cells and GECs.12, 18
P. gingivalis, a major etiological agent of chronic periodontal diseases, produces a broad array of potential virulence factors: lipopolysaccharide, fimbriae, hemagglutinin, hemolysins, and gingipains.32 These virulence factors could trigger GECs through pathogen-associated molecular pattern receptors such as TLR2/TLR433 or protease-activated receptor 2,34 which lead to the activation of transcription signaling pathways NF-κB, MAPK (mitogen-activated protein kinase), and MEK/ERK (mitogen-activated protein kinase kinase/extracellular regulated kinase).14, 35 It is becoming apparent that both recruitment of NF-κB to target promoters and NF-κB-induced transcriptional genes are modulated through chromatin modification.36 The decrease of DNMT has been reported to be associated with impaired signaling through the ERK and JNK (c-Jun N-terminal kinase) pathways.37 It is therefore likely that connections exist between these transduction signaling pathways and DNA methyltransferase and histone deacetylase. On the other hand, our gene network analysis depicted the relationships between the queried genes and other immune response genes in the same network. Innate immune markers DEFB4 (hBD2), CCL20, DEFB1 (hBD1), DEFB103A (hBD3), and IL8 show intermediate connections to methylated genes tested, and also show direct or indirect connection with the genes NF-κB (MYD88), TLR signaling (TLR2 and TLR4), and JAK/STAT (STAT5, STA5A, STAT5B, JAK2, and GATA1). In addition, several other cytokines (IL-6, IL-1B, and IL-4) and chemokines (CCL19, CCL21, CCR7, and CCL2) are also involved in this network, which are importantly relevant for immunoregulation. Therefore, it appears that the possible interaction between innate immune responses and epigenetic events, and epigenetic changes might determine the “on” or “off” state of transcriptional activity of signaling transduction pathways. However, the exact signal triggering epigenetic changes remains unclear.
It has been well known that many factors have contributed to the pathogenesis of periodontal disease. Genetic factors have a strong impact on the pathogenesis of disease developments; i.e., gene polymorphism of TLR is associated with susceptibility to chronic periodontitis.38 However, increasing evidences have shown that the factors that influence the epigenome during the lifetime may also change the host response to environmental exposures, such as bacterial infection. It was reported that inflammatory signals that activate NF-κB have been shown to be involved in histone methylation and acetylation, resulting in the selective gene induction.39 Epigenetic modifications alter patterns of gene expression, which in turn leads to various clinical outcomes. For example, the alteration in DNA methylation status of the Igf2 gene in murine placental tissues was associated with maternal infection with the periopathogenic bacteria Campylobacter rectus.10 This may be particularly relevant in the disease development of humans, as C. rectus exposure of the fetus has been found to be associated with preterm delivery,40 and impaired fetal growth has been associated with adult-onset diabetes and cardiovascular disease.41 Furthermore, variations in epigenetic status will likely elicit diverse inflammatory responses in periodontal disease. An earlier study reported that epigenetic mechanisms have a major role in the inflammation processes, locally as well as systemically.42 Prostaglandin-endoperoxide synthase-2 promoter methylation levels from periodontally inflamed gingival biopsies showed a significant increase when compared with noninflamed biopsy samples,43 suggesting that methylation status is associated with chronic inflammation. Andia et al.44 reported that hypomethylated status of IL-8 gene promoter was associated with aggressive periodontitis. Taken together, the epigenetic modification would have a high impact on disease developments, and it would be of interest to understand how epigenetic mechanism contribute to the pathogenesis of inflammation, and how local persistent chronic inflammation and systemic disease are connected.45
Periodontitis is not just chronic inflammatory disease, but its persistent nature could also exert a significant systemic impact on health, such as atherosclerosis, aspiration pneumonia, diabetes, adverse pregnancy outcomes, and rheumatoid arthritis.46, 47, 48, 49 Although the strategies for prevention and therapies for the periodontal disease have improved recently, effective prevention and treatments are still a challenge. It is well known that the pathogenesis of periodontal disease is initiated by bacteria populating the tooth-associated biofilm, but disruption of host innate immune response may be a major contributory factor for disease progression.50 The traditional treatment for periodontal disease with direct targeting of pathogen or inflammation has limitations, as many treated patients develop recurrent disease and bacterial resistance. Thus, developing new therapeutic strategy for chronic inflammation by inducing host innate immune response is highly desirable.
Various HDAC inhibitors and DNMT inhibitors have already been evaluated as therapeutic agents with the potential roles in cancer treatments.2 Recently, agents derived from those inhibitors have been shown to have effective therapeutic roles beyond cancer treatments, based on their anti-inflammatory properties.51 For example, SB has been used to stimulate production of endogenous antimicrobials in the gut, which can promote the clearance of Shigella, leading to clinical improvement of chronic infection. In this study, we have found that the inhibition of chromatin-modifying enzymes HDAC and DNMT have varying degrees of roles in the induction of antimicrobial function in GECs in response to various oral bacteria, suggesting the potential role of immune therapy of these inhibitors through epigenetic modification in oral cavity.
In this study we report for the first time how epigenetic mechanism has an important role in the expression of antimicrobial molecules in bacteria–host cell interactions. In summary, our data identified several potentially important effects of oral bacteria on epigenetic state of GECs and provide new insights that are critical for innate immune and inflammatory responses. We anticipate that the work of this study will lead to new understanding of coordinated interplay between epigenetic mechanisms, gene expression, and environment that can be translated into development of innovative treatments targeted for periodontitis.
Methods
Reagents. The inhibitors for histone deacetylase, SB (0.5 and 2.0 mm) and TSA (0.5 and 9 nm), and for DNA methylation, AZA (1 and 10 μm), were from Sigma-Aldrich (St Louis, MO). Primary antibodies used were anti-DNMT1, anti-HDAC1, HDAC2 (mouse monoclonal antibody; Active Motif, Carlsbad, CA), and anti-actin (mouse monoclonal; Active Motif). Secondary antibody was anti-mouse IgG horseradish peroxidase-conjugated antibody from Jackson Immune Research (West Grove, PA).
Human primary GECs, human immortalized keratinocytes (TERT), and bacteria culture. Gingival biopsies were obtained from healthy patients who underwent third-molar extraction in the Department of Oral Surgery, School of Dentistry, University of Washington, in accordance with the University of Washington institutional review board-approved study. Epithelial cells were cultured in keratinocyte growth medium with 0.15 mm Ca2+ using the supplements from the keratinocyte growth medium-bullet kit (Cambrex, Walkersville, MD) at 37 °C in a humidified atmosphere containing 5% CO2.
TERT cells were cultured in serum-free medium (GIBCO/Invitrogen, Carlsbad, CA), supplemented as described previously52 with 25 μg ml–1 bovine pituitary extract, 0.2 ng ml–1 epidermal growth factor, and 0.4 mmol l–1 CaCl2.
P. gingivalis (ATCC (American Type Culture Collection, Manassas, VA) 33277) was cultured under anaerobic conditions (85% N2, 10% H2, and 5% CO2) at 37 °C in Trypticase soy broth (BBL, Sparks, MD) supplemented with 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione per liter. F. nucleatum (ATCC 25586) was grown in Todd-Hewitt broth supplemented with 1 g of yeast extract per 100 ml at 37 °C under anaerobic conditions. Bacterial purity was determined by Gram staining, and numbers were estimated by absorbance measurement using the TECAN GENios Multidetection Reader, V.4.51 (Phoenix, Hayward, CA). The bacteria were cultured to late logarithmic growth phase. GECs and TERT cells were grown to 85% confluence and stimulated with various stimuli as indicated. Bacteria were used at an amount equivalent to the indicated MOI. Each stimulation experiment was performed in duplicate, and cells from two to four different donors were tested.
QRT-PCR. Total RNA was isolated from GECs using RNeasy mini kit (Qiagen, Valencia, CA). Complementary DNA was prepared from 0.5 μg total RNA. Quantitative analysis of the complementary DNA was performed using an iCycler iQ5 (Bio-Rad, Hercules, CA). QRT-PCR was conducted with SYBR Green Supermix kit from Bio-Rad. Melt-curve analysis was performed to confirm that the signal was of the expected amplification product. The sequences of oligonucleotide primers for hBD2 and CCL20 were described previously (Yin and Dale21). The sequences of HDAC1, HDAC2 and DNMT1 are: HDAC1 forward sequence 5′-CATCTCCTCAGCATTGGCTT-3′, HDAC1 reverse sequence 5′-CGAATCCGCATGACTCATAA-3′, HDAC2 forward sequence 5′-ATGAGGCTTCATGGGATGAC-3′, HDCA2 reverse sequence 5′-ATGGCGTACAGTCAAGGAGG-3′, DNMT1 forward sequence 5′-CCTGTACCGAGTTGGTGATGGT-3′, DNMT1 reverse sequence 5′-CCTTCCGTGGGCGTTTC-3′, GAPDH (glyceraldehydes-3-phosphate dehydrogenase) forward sequence 5′-AATGAAGGGGTCATTGATGG-3′, and GAPDH reverse sequence 5′-AAGGTGAAGGTCGGAGTCAA-3′. Each sample was normalized to the level of GAPDH transcript. The calculation for relative fold changes was described previously.21
Cytokine secretion assay. Cell culture supernatants after various stimulations were collected, and the level of IL-8 secretion was measured with an enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN) in accordance with the manufacturer's protocol. A standard curve was generated with each set of samples assayed. The optical density value of blank control (reagent diluents) was subtracted from the measured optical density of the different standards and samples. Final concentrations in each sample were calculated as the mean of the results at the proper sample dilution yielding optical densities in the linear parts of the standard curves. Supernatants of cells from three different donors were tested, with duplicate samples from each donor.
Western blotting. Protein levels of DNMT1, HDAC1, and HDAC2 in GECs were assessed by western blot analyses. Briefly, GECs were treated with various bacterial stimulations (MOI 100:1), and nuclear extracts were prepared using Nuclear Extraction Kit from Active Motif. Nuclear protein concentration was determined using bicinchoninic protein assay kit (Thermo Fisher Scientific, Rockford, IL), and samples were resolved on pre-cast NuPAGE Bis-Tris 10–12% denaturing polyacrylamide gels (Invitrogen, Carlsbad, CA), which were transferred to methanol-activated Immobilon-P polyvinylidene difluoride membranes (Millipore, Billerica, MA) and probed with specific primary antibodies. Nuclear extracts of HeLa cells (Active Motif) were used as positive controls. The signals were detected by chemiluminescence (Thermo Fisher Scientific) and exposure to X-ray film (Thermo Scientific, Rockford, IL).
Analysis of DNA methylation. Genomic DNA was prepared from human TERT cells using Perfect Pure DNA Cultured Cell kit from 5 Prime (Gaithersburg, MD) according to the manufacturer's instruction. To analyze DNA methylation of PCR array, 0.5 μg genomic DNA was performed for the digestion of unmethylated and methylated DNA, using methylation-sensitive and methylation-dependent restriction enzymes. The remaining DNA after digestion is quantified by real-time RT-PCR, using primers that specifically flank the region of interest (Human Cytokine production, Methyl-Profile PCR Arrays; SABiosciences, Frederick, MD). PCR array plates were processed in a Bio-Rad iCyler Real-Time PCR System, using automated baseline and threshold cycle detection. Data were analyzed using the Excel-based PCR array data analysis template from SABiosciences. For this analysis, the relative concentrations of differentially methylated DNA (specifically, hypermethylated, unmethylated, and intermediary methylated DNA) are determined by comparing the amount of each digest with that of a mock digest. The data are expressed as the mean of the percent hyper-methylated DNA. All our experiments were repeated twice using TERT cells.
Tri-methyl histone H3 (lys4) protein measurement. GECs were treated as indicated previously, and the cells were washed with cold phosphate-buffered saline and harvested using cell lysis buffer including fresh phenylmethylsulfonyl fluoride (Cell Signaling, Danvers, MA). Cell lysates were prepared followed by brief sonication. The levels of tri-methyl histone (lys4) proteins were measured with PathScan (Cell Signaling) enzyme-linked immunosorbent assay in accordance with the manufacturer's protocol.
Gene networks analysis. PubGene Bio-networks (http://www.pubgene.com/public.html) were used to search for literature gene networks.53 This program provides biomedical knowledge from publicly available gene and databases, and creates a gene-to-gene co-citation network.
Data analysis. Each experiment was performed with cells from at least two to three different donors, and within an experiment, each test condition was performed in duplicate. Values are shown as the mean±s.e.m. from multiple experiments. Statistical significance was determined using one-way analysis of variance (JMP for Windows Release 6.0; SAS Institute, Cary, NC) among the groups followed by the two-tailed Student's t-test (Excel Window 2007, Seattle, WA). Differences were considered to be statistically significant at the cutoff level of P<0.05.
Accession codes
References
Rodenhiser, D. & Mann, M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ 174, 341–348 (2006).
Egger, G., Liang, G., Aparicio, A. & Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Socransky, S.S., Haffajee, A.D., Cugini, M.A., Smith, C. & Kent, R.L. Jr . Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998).
Kinane, D.F. & Hart, T.C. Genes and gene polymorphisms associated with periodontal disease. Crit. Rev. Oral. Biol. Med. 14, 430–449 (2003).
Michalowicz, B.S. et al. Evidence of a substantial genetic basis for risk of adult periodontitis. J. Periodontol. 71, 1699–1707 (2000).
Feinberg, A.P. Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–440 (2007).
Schenkein, H.A. Finding genetic risk factors for periodontal diseases: is the climb worth the view? Periodontol. 2000 30, 79–90 (2002).
Offenbacher, S., Barros, S.P. & Beck, J.D. Rethinking periodontal inflammation. J. Periodontol. 79, 1577–1584 (2008).
Bobetsis, Y.A. et al. Bacterial infection promotes DNA hypermethylation. J. Dent. Res. 86, 169–174 (2007).
Hancock, R.E. Peptide antibiotics. Lancet 349, 418–422 (1997).
Yin, L. et al. Differential and coordinated expression of defensins and cytokines by gingival epithelial cells and dendritic cells in response to oral bacteria. BMC Immunol. 11, 37 (2010).
Chung, W.O. & Dale, B.A. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72, 352–358 (2004).
Chung, W.O. & Dale, B.A. Differential utilization of NFκB signaling pathways for gingival epithelial cell responses to oral commensal and pathogenic bacteria. Oral Microbiol. Immunol. 23, 119–126 (2008).
Joly, S., Maze, C., McCray, P.B. Jr . & Guthmiller, J.M. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42, 1024–1029 (2004).
Yang, D. et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74, 448–455 (2003).
Dommisch, H. et al. Protease-activated receptor 2 mediates human beta-defensin 2 and CC chemokine ligand 20 mRNA expression in response to proteases secreted by Porphyromonas gingivalis. Infect. Immun. 75, 4326–4333 (2007).
Chung, W.O. et al. Interplay of protease-activated receptors and NOD pattern recognition receptors in epithelial innate immune responses to bacteria. Immunol. Lett. 131, 113–119 (2010).
Yan, C. & Boyd, D.D. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol. Cell Biol. 26, 6357–6371 (2006).
Simon, R. et al. Analysis of gene expression data using BRB-array tools. Cancer Inform. 3, 11–17 (2007).
Yin, L. & Dale, B.A. Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human beta-defensin-2. J. Med. Microbiol. 56, 976–987 (2007).
Barrett, T. et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 35, D760–D765 (2007).
Hasegawa, Y. et al. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect. Immun. 75, 2540–2547 (2007).
Maekita, T. et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin. Cancer Res. 12, 989–995 (2006).
Dai, K.S. & Liew, C.C. Chromosomal, in silico and in vitro expression analysis of cardiovascular-based genes encoding zinc finger proteins. J. Mol. Cell Cardiol. 31, 1749–1769 (1999).
Persson, G.R. & Persson, R.E. Cardiovascular disease and periodontitis: an update on the associations and risk. J. Clin. Periodontol. 35, 362–379 (2008).
Schulze-Luehrmann, J. & Ghosh, S. Antigen-receptor signaling to nuclear factor kappa B. Immunity 25, 701–715 (2006).
Rhee, I. et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404, 1003–1007 (2000).
Jones, P.A. & Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 (2002).
Starner, T.D., Barker, C.K., Jia, H.P., Kang, Y. & McCray, P.B. Jr . CCL20 is an inducible product of human airway epithelia with innate immune properties. Am. J. Respir. Cell Mol. Biol. 29, 627–633 (2003).
Lamont, R.J. & Jenkinson, H.F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62, 1244–1263 (1998).
Holt, S.C., Kesavalu, L., Walker, S. & Genco, C.A. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20, 168–238 (1999).
Darveau, R.P. et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72, 5041–5051 (2004).
Chung, W.O., Hansen, S.R., Rao, D. & Dale, B.A. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 173, 5165–5170 (2004).
Huang, G.T., Zhang, H.B., Dang, H.N. & Haake, S.K. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 37, 303–312 (2004).
Natoli, G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb. Perspect. Biol. 1, a000224 (2009).
Chen, Y., Gorelik, G.J., Strickland, F.M. & Richardson, B.C. Decreased ERK and JNK signaling contribute to gene overexpression in “senescent” CD4+CD28− T cells through epigenetic mechanisms. J. Leukoc. Biol. 87, 137–145 (2010).
Noack, B., Gorgens, H., Lorenz, K., Schackert, H.K. & Hoffmann, T. TLR4 and IL-18 gene variants in chronic periodontitis: impact on disease susceptibility and severity. Immunol. Invest. 38, 297–310 (2009).
Ito, K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem. Soc. Trans. 35, 281–283 (2007).
Madianos, P.N. et al. Maternal periodontitis and prematurity. Part II: maternal infection and fetal exposure. Ann. Periodontol. 6, 175–182 (2001).
Barker, D.J., Eriksson, J.G., Forsen, T. & Osmond, C. Fetal origins of adult disease: strength of effects and biological basis. Int. J. Epidemiol. 31, 1235–1239 (2002).
El Gazzar, M., Yoza, B.K., Hu, J.Y., Cousart, S.L. & McCall, C.E. Epigenetic silencing of tumor necrosis factor alpha during endotoxin tolerance. J. Biol. Chem. 282, 26857–26864 (2007).
Zhang, S. et al. Alteration of PTGS2 promoter methylation in chronic periodontitis. J. Dent. Res. 89, 133–137 (2010).
Andia, D.C. et al. DNA methylation status of the IL8 gene promoter in aggressive periodontitis. J. Periodontol. 81, 1336–1341 (2010).
Wilson, A.G. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J. Periodontol. 79, 1514–1519 (2008).
Xiong, X., Buekens, P., Fraser, W.D., Beck, J. & Offenbacher, S. Periodontal disease and adverse pregnancy outcomes: a systematic review. BJOG 113, 135–143 (2006).
Awano, S. et al. Oral health and mortality risk from pneumonia in the elderly. J. Dent. Res. 87, 334–339 (2008).
de Pablo, P., Chapple, I.L., Buckley, C.D. & Dietrich, T. Periodontitis in systemic rheumatic diseases. Nat. Rev. 5, 218–224 (2009).
Pihlstrom, B.L., Michalowicz, B.S. & Johnson, N.W. Periodontal diseases. Lancet 366, 1809–1820 (2005).
Darveau, R.P. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490 (2010).
Halili, M.A., Andrews, M.R., Sweet, M.J. & Fairlie, D.P. Histone deacetylase inhibitors in inflammatory disease. Curr. Top Med. Chem. 9, 309–319 (2009).
Rheinwald, J.G. et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol. Cell Biol. 22, 5157–5172 (2002).
Jenssen, T.K., Laegreid, A., Komorowski, J. & Hovig, E. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 28, 21–28 (2001).
Acknowledgements
We thank Mr Dennis DiJulio, Mr Jonathan An, and Miss Julia Tracy for technical assistance. This study was supported by the NIH/NIDCR grants RO1 DE 013573 and DE 19632.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yin, L., Chung, W. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol 4, 409–419 (2011). https://doi.org/10.1038/mi.2010.83
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2010.83