Abstract
Systemic immunization of macaques with a combination of DNA-poxvirus-based vaccines confers protection from high level of both systemic and mucosal viral replication following rectal exposure to the pathogenic SIVmac251. Here we investigated early post-infection events in rectal and vaginal tissues, and found that the loss of CCR5+CD4+ T cells was equivalent in vaccinated and control macaques, despite a three logs reduction at mucosal sites of simian immunodeficiency virus (SIV) RNA in the vaccinated group. Even though a normal CD4+ T cell number is not reconstituted at mucosal sites in either group, vaccination appeared to confer a better preservation of the CD4+ CCR5+ T cells that replenish these sites. Analysis of rectal tissues RNA following challenge exposure demonstrated a decreased expression in vaccinated macaques of transforming growth factor-β, cytotoxic T lymphocyte antigen-4, FoxP3, and indoleamine 2,3-dioxygenase, an immune suppressive enzyme expressed by dendritic cells that converts tryptophan to kynurenine and limits T-cell responses. Accordingly, the ratio of kynurenine and tryptophan in the plasma was significantly reduced in the vaccinated animals respect to the controls. Thus, preexisting adaptive immune responses induced by these vaccine modalities, although they do not protect from CD4+ T-cell depletion, nevertheless, they contain SIVmac251 replication and delay expression of markers of T-cell activation and/or suppression at mucosal sites.
Similar content being viewed by others
Introduction
Untreated infections of humans with the immunodeficiency virus (HIV) and of macaques with pathogenic strains of simian immunodeficiency virus (SIV) result in loss of the memory CD4+ T-cell population at mucosal sites within the first weeks from the infection.1, 2, 3 and 4 This early depletion of CD4+ T cells occurs more noticeably in the gut1, 2 because this compartment is enriched of activated CD4 population that express the CCR5 receptor and are targets for the infection.5, 6 and 7 It is believed that CD4+ T-cell depletion at mucosal sites is central to AIDS pathogenesis.2, 3 and 8 Indeed, animals that show a better capacity to repopulate the gut with CD4+ T cells appear to progress more slowly to AIDS9 and animals treated during primary SIV infection with antiretroviral therapy maintain higher levels of CD4+ T cells at mucosal sites,10, 11 suggesting that early inhibition of viral replication decreases the severity of CD4+ T-cell loss. Interestingly however, studies on a non-pathogenic natural model of SIV infection of sooty mangabey indicate that early CD4+ T-cell loss “per se”, whereas perhaps necessary, is not sufficient for disease development.12, 13 In fact in this model, SIV infection results in severe loss of CD4+ T cells at mucosal sites, yet these animals do not develop disease.13
The finding of high levels of immune activation in pathogenic models of SIV and in HIV infections, but not in SIV infection of sooty mangabey, has led to the hypothesis that immune activation is key to AIDS pathogenesis.14 The increased turnover of T cells observed in HIV-infected individuals and in pathogenic models of SIV infection of macaques is likely due to the homeostatic response to the initial loss of cells that, together with high level of immune activation, may drive the progressive decline of the CD4 T-cell number.14, 15 and 16 Indeed, in the non-pathogenic SIV model, the level of immune activation is considerably lower. Thus, tempering the loss of the CD4+ T cells and lowering immune activation may delay progression to AIDS.
Systemic immunization of macaques with live vector-based SIV vaccines has resulted in the elicitation of both systemic and mucosal CD8+ and CD4+ T-cell responses that are recalled following exposure to SIV.17, 18, 19, 20 and 21 The frequency of these responses has been linked to decreased viral replication in some, if not all, of these studies. Indeed, several vaccines based on the combination of DNA followed by poxvirus-based SIV vaccines are able to reduce SIVmac251 plasma and tissue and to delay progression to disease.17, 18, 19, 20, 21 and 22 However, whether this reduction in virus levels is sufficient to protect the mucosal compartment from CD4+ T-cell loss and whether vaccination affects immune activation have not yet been thoroughly investigated.
The necessity to study whether these vaccine modalities protect from CD4+ T-cell loss rises because early depletion of helper CD4+ T cells may affect long-term CD8 and B-cell responses to the virus23, 24, 25 and 26 and abrogate the long-term effect of vaccination. Indeed, experimental depletion of CD4+ T cells at the time of infection decreases the durability of humoral and T-cell responses.27 Similarly, vaccination under conditions of CD4+ T-cell deficiency decreases the titers of antibodies to SIV, the quality of CD8+ T-cell responses, and the durability of protection following challenge exposure to SIVmac251.27
A report has analyzed in detail whether vaccination protects the gut from destruction of mucosal CD4+ T cells following SIVmac251 challenge exposure.28, 29 In that study, a DNA prime/recombinant adenovirus 5 SIV boost vaccine regimen decreased virus levels by approximately one log in plasma and tissues of the vaccinated macaques.29 The authors found modest differences in the ratio of naive and memory CD4+ T cells in tissues of vaccinated and naive control animals.
In the present study, we addressed this question by obtaining serial blood and mucosal samples from DNA/modified vaccinia Ankara (MVA)-SIV-vaccinated macaques in the acute and chronic phases of infection. We demonstrated that despite the ability of systemic vaccination to significantly decrease mucosal virus levels, the loss of CD4+ T cells at mucosal sites did not differ from that of non-vaccinated controls. Rather, we observed that a reduced virus level at mucosal sites was associated with lower expression of molecules that modulate immune responses, such as cytotoxic T lymphocyte antigen-4 (CTLA-4), FoxP3, and indoleamine 2,3-dioxygenase (IDO).
Results
Vaccination with DNA/MVA-SIV induces immune responses and significantly decreases virus levels in tissues following challenge exposure to SIVmac251
Macaques were vaccinated with the re-assorted SIV-Retanef gene and the SIV gag, pol, and env genes expressed in different plasmids (DNA-SIV-gpe)30, 31 at weeks 0 and 4, followed by two boosts at week 26 and at week 53, with recombinant MVA expressing the same antigens. A group of six macaques were mock vaccinated with a nonrecombinant MVA vector as controls (Figure 1a).
Study design and simian immunodeficiency virus (SIV)-specific immune responses elicited by vaccination. (a) Immunization regimens. The long arrows represent vaccination and challenge time. The short arrows represent time of collection of rectal and vaginal pinches biopsies. Mamu-A*01+ animals in each group are identified with bold character. (b) p27 Gag and gp120 Env lymphoproliferative responses following MVA-SIV immunization (see (a) and arrows) of vaccinated macaques. The lines represent the average of the seven vaccinated animals. Intracellular cytokine staining on peripheral blood mononuclear cells (PBMCs) at week 55 (c) and 72 (d, e). Bars represent the average frequency of CD8+ and CD4+ T cells that produce interferon (IFN)-γ or interleukin (IL)-2. Values are expressed as percentage of cytokine-positive cells with background subtracted. (f) Immune response to SIV Gag measured by IFN-γ ELISPOT assay in fresh PBMCs following a stimulation with overlapping Gag and Env peptide pools. Data are expressed as number of spots per 106 cells. The line represents the average and standard deviation for all the seven vaccinated (VACC) animals. (g) Frequency Gag181 tetramer+ CD8+ T-memory cells (line) central memory (CM=dark bars) or effectors (white bars) in blood. Line and bars represent the average±the standard deviation of the three Mamu-A*01+ animals in the vaccinated (VACC) group. (h) Expansion of central memory (TCM) Gag181 tetramer+ CD8+ T cells following infection with SIVmac251 in the blood of vaccinated (n=3) and mock-vaccinated (n=2) macaques. TCM are defined as CD28+/CD95+ CM9+ CD8 T cells. Bars represent the average of the frequency of TCM within the tetramer-positive population±standard deviation. (i) Staining of purified mononuclear cells obtained from rectal mucosa biopsies with the CM9 tetramer. Raw data on central memory and effector memory CM9+ T cells in two vaccinated and two control macaques. Mean values of total memory CM9+ cells obtained from vaccinated macaques 902L, 906L and 314M and from control animals 224M and 300M (j). (k) Intracellular cytokine staining. Bars represent the average of the frequency of IL-2-producing CD8+ T cells following in vitro stimulation with gag, env, pol peptides in six vaccinated animals and four mock-vaccinated animals, at week 12 (left panel) and week 16 (right panel) after infection. For each animal the background frequency of cytokine production was subtracted from the positive samples prior calculation of the average.
The immunogenicity of the same DNAs expressed in the context of the NYVAC vector in naive macaques has been demonstrated previously.31 To validate the immunogenicity of these new MVA recombinant viruses, we measured the lymphoproliferative responses to Gag p27 and to Env gp120 in the blood after immunizations with recombinant MVA-SIV. The peak of proliferative responses to both proteins was observed 1 week after the first immunization, as expected with these vaccine modalities, and this response contracted by week 28 (Figure 1b). The second immunization with MVA did not further boost these proliferative responses as also observed previously with similar vaccine modalities.31 Vaccinated animals had measurable CD8+ and CD4+ T-cell responses in peripheral blood mononuclear cells at week 55 measured after stimulation with SIV- Gag-, Pol-, and Env-overlapping peptides (Figure 1c).
The CD4+ and CD8+ T-cell responses were detectable up to week 72, 2 weeks before challenge exposure to SIVmac251 (Figure 1d and e). Accordingly, interferon (IFN)-γ enzyme-linked immunosorbent spot (ELISPOT) were measurable during the all immunization interval (Figure 1f). DNA vaccination induced low levels of SIV Gag-specific CD8+ T-cell responses in Mamu-A*01+ animals measured with the CM9 tetramer, which were boosted effectively after the first MVA immunization but not after the second immunization (Figure 1g). Low frequency of SIV Tat-specific CD8+ T cells, measured with the Tat-SL8 tetramer in Mamu-A*01+ macaques, was elicited in all vaccinated macaques (data not shown). As expected, CM9 or SL8 tetramers failed to stain cells from the mock-immunized macaques (data not shown).
Analysis of the frequency of central memory and effector memory T cells within the CM9 tetramer+ population in blood and rectal mucosa was performed using the CD95 and the CD28 markers32 following exposure to SIVmac251. We found a significantly better expansion of central memory: CD95+ CD28+ CM9 tetramer+ in vaccinated than mock-vaccinated macaques in blood (Figure 1h). Similarly a better expansion of tetramer+ was found in the rectal mucosa of vaccinated animals (Figure 1i and j). Accordingly, a trend for a higher frequency of IL-2-producing CD8+ T cells was also found in the blood of the six vaccinated animals following stimulation with Gag, Env, or Pol peptides at later time points (weeks 12 and 16) than in four mock-vaccinated animals analyzed as controls33 (Figure 1k).
All macaques were intrarectally exposed to SIVmac251 at week 74 (see Figure 1a), as previously described.18 As expected, this vaccination regimen reduced plasma virus levels of 100-fold during both primary as well as during chronic infection (Figure 2a–c). In fact, plasma viral load on vaccinated animals was significantly lower then controls at weeks 2, 12, and 16 after the SIV exposure, whereas the difference in plasma virus level between immunized and control animals became nonsignificant at week 24 (Figure 2c).
Virological outcome following intrarectal challenge exposure to SIVmac251. (a) Plasma virus level in each macaque from the vaccinated group (VACC) and (b) mock-vaccinated group (MOCK). Mamu-A*01+ animals in each group are identified with bold character in the legend and with red symbols and lines in the graphs. (c) Geometrical mean virus load in vaccinated and mock-vaccinated group. Bars represent standard deviation. The P-values refer to weeks 2, 12, 16, and 24 after challenge. (d) CD4 mRNA expression and simian immunodeficiency virus (SIV) Gag RNA copies in mononuclear cells from axillary lymph nodes of vaccinated (n=5; 902L, 906L, 306M, 318M, 307M) and mock-vaccinated animals (n=5; 224M, 300M, 552M, 556M, 557M) at week 24 post-infection. Data are represented as average values plus standard deviations. (e) SIV-RNA Gag copies for microgram of total RNA in rectal biopsies (f) or vaginal biopsies collected from the seven vaccinated and the five mock-vaccinated animals during the course of chronic infection, at weeks 12, 16, and 24 weeks post-challenge. The horizontal bars represent the average value in each group.
A decrease in virus levels that approached statistical significance was also observed in the lymph nodes (LNs) of the vaccinated macaques (Figure 2d). Analysis of virus levels at mucosal sites was performed at weeks 12, 16, and 24 after infection in both rectal and vaginal biopsies. Interestingly, this analysis revealed a decrease of 1,000-fold in virus levels at both mucosal sites of vaccinated macaques (Figure 2e and f). Thus, systemic immunization was able to reduce plasma virus levels and, surprisingly, this reduction was even more evident at mucosal sites.
Vaccination does not protect mucosal sites from early CD4+ T-cell loss
Having obtained evidence that vaccination was associated with a decrease of three and two logs of virus levels at mucosal sites and in plasma, we wished to evaluate whether prior vaccination spared CD4+ T cells. Serial blood and mucosal samples were collected from all animals 2 weeks before challenge exposure (week 72), in the early phase of infection (2 weeks after infection) and during chronic infection at weeks 12, 16, and 24 and mononuclear cell preparations were stained for CD4, CD8, CCR5, or CD95, and CD45RA. An example of raw data is presented in Figure 3a.
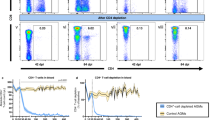
CD4+ T cells in blood and tissues of vaccinated and mock-vaccinated animals. (a) Activated CD4+ T cells were defined by gating on the lymphocyte population positive for CD3. The frequency of CCR5+/CD4+ cells was then calculated. The density plot represents mononuclear cells extracted from a vaginal mucosa of a naive noninfected animal. CD4 T cells were gated in the lymphocyte population and naive CD4+ T cells were defined as CD45+ CD95− T cells, whereas memory T cells were gated as CD95+/CD45RA+ CD45− T cells. (b) Frequency of CCR5+ CD4+ T cell in the blood of vaccinated (left) and mock-vaccinated (right) animals. Lines represent the mean values. (c) Absolute number of memory (CD95+/CD45RA+ or CD45RA) and (d) naive (CD95−/CD45RA+), CD3+/CD4+ T cells in the blood. Bars represent the average values for each group±standard deviation. White bars represent pre-challenge levels at week 72. (e) Frequency CD4+ CD95+ cells and (f) activated CCR5+ CD4+ T cells CD3+/CD4+/CCR5+) in the rectal mucosa of vaccinated and mock-vaccinated animals. Bars represent the average values for each group±standard deviation. The white bars represent the pre-challenge basal level (week 72).
Prior SIV infection, the absolute number of the total memory and naive CD4+ T cells, defined with CD95 and CD45RA markers or the absolute number of activated CD4+ T cells (CCR5+/CD4+), did not differ in the blood of vaccinated and mock-vaccinated animals (data not shown) therefore, the data from the animals studied were grouped to calculate pre-challenge baseline values. In blood, the frequency of the total population of CD3+/CD4+ cells and of activated CCR5+ CD4+ T cells did not differ significantly between the control and vaccinated groups before and after challenge exposure (data not shown; Figure 3b). Overall, no durable differences in the absolute number of memory CD95+ CD45RA+ CD4+ or naive CD95− CD45RA+ CD4+ was observed after challenge exposure in the blood of the vaccinated vs. mock-vaccinated animals (Figure 3c and d).
At mucosal sites, the frequency of CD4+ T cells was assessed 2 weeks before challenge, and at 2−3, 12, 16, and 24 weeks from challenge exposure. No significant differences were observed in the frequency of memory CD95+/CD4+ T cells within few weeks from infection in rectal and vaginal mucosa (Figure 3e; data not shown), suggesting that the loss of the memory CD4+ population during the acute phase of infection occurs to a similar extent in the mucosal compartment of vaccinated and control macaques. In rectal mucosa, a similar loss of CCR5+ CD4+ T cells occurred in vaccinated and mock-vaccinated animals at 2 or 3 weeks from challenge exposure. However, we observed an apparently better preservation of the CCR5+ CD4+ T cells that repopulate this site in the vaccinated animals (Figure 3f), suggesting that prior vaccination may protect newly recruited CD4+ T cells at mucosal sites.
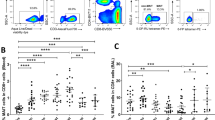
Because most of the data described above were derived from mononuclear cells suspension of minced mucosal tissues, we wanted to confirm our observations by direct enumeration of CD4+ T cells in intact tissues. To this aim, we enumerated by immune histochemistry CD4+ T cells expressing memory and activation markers in rectal pinch biopsies collected before and after SIVmac251 challenge exposure. An example of raw data is presented in Figure 4a. By 2 weeks from infection, the number of CD4+/mm2 of rectal tissue was decreased of two-thirds as compared to the pre-challenge levels in both vaccinated and mock-vaccinated macaques and normal levels were not reconstituted thereafter (Figure 4b). In contrast, but not unexpectedly, we observed an increase in the absolute number of CD8+ cells in the same tissue from both groups starting 2 weeks following challenge exposure (week 76; Figure 4c).
CD4+ T-cell subsets in rectal mucosa of vaccinated and mock-vaccinated animals. (a) Representative high-power (× 40) fields of lamina propria from rectal sections from one vaccinated (902L) and one mock-vaccinated animal (300M) at 2 weeks prior infection (−2 or week 72) and 2 and 16 weeks after challenge with SIVmac251. In situ detection of CD4+/CCR5+ cells (left) and of CD4+/Ki67+ cells (right). The bar corresponds to 25 μm. (b) Absolute number of CD4+ cells/mm2 and (c) CD8+ T cells/mm2 in intact rectal mucosa pinches. (d) Absolute number (cell/mm2) of CD4 expressing CD95 or (e) CCR5 or (f) Ki67 in intact rectal mucosa pinches collected from vaccinated and mock-vaccinated animals. Bars represent the average values at each time point±standard deviation.
Enumeration in rectal biopsies of the absolute number of CD95+/CD4+ cells also demonstrated a decrease of two-thirds of memory CD4+ T cells in both the groups of animals with respect to the pre-challenge levels, and normal levels of these cells were not reconstituted overtime (Figure 4d). Analysis of the number of CCR5+/CD4+ T cells/mm2 of tissues revealed only a trend toward higher numbers in the tissue of vaccinated macaques a few weeks after challenge exposure, but the overall difference did not reach statistical significance (Figure 4e). Similarly, no significant differences were observed in the number of the CD4+ Ki67+ T-cell subset in tissues following SIVmac251 infection (Figure 4f). A parallel analysis of LNs also revealed no significant differences in number of CD3+ CD4+ and CD4+ Ki67+ cells in vaccinated or mock-vaccinated macaques (data not shown).
Vaccination delays expression of modulators of immune responses at mucosal sites
The results presented above indicate that although vaccination does not protect from CD4+ T-cell loss it does protect, at least transiently, from high virus levels at the mucosal site. Recent work has suggested an increased frequency of CD25/FoxP3/CTLA-4 CD4+ T cells, transforming growth factor (TGF)-β, and in the tissues of SIV-infected macaques.34, 35 Because the expression of these molecules may be associated with suppression of SIV-specific immune responses and be detrimental to the host ability to control viral replication,36 we investigated whether preexisting adaptive responses to SIV modulated the expression of these immune modulators.37 The level of mRNAs for TGF-β, CTLA-4, FoxP3, and IDO was measured by reverse transcription–PCR in the total RNA extracted from rectal and vaginal pinch biopsies of vaccinated and mock-vaccinated macaques after challenge exposure to SIVmac251. We observed that the expression of TGF-β, CTLA-4, and FoxP3 mRNAs was upregulated at an earlier time in mock-vaccinated compared to vaccinated macaques in both rectal (Figure 5a–d) and vaginal tissues during chronic infection (data not shown). Furthermore, the expression of CTLA-4 and FoxP3 was correlated in the mock-vaccinated group (P<0.1, R=0.94) but not in the vaccinated animals (P=0.87, R=0.04) (Figure 5e and f).
Expression of activation/immune suppression markers in rectal biopsies. (a) Quantification of the expression of mRNAs for transforming growth factor (TGF)-β, (b) cytotoxic T lymphocyte antigen-4 (CTLA-4), (c) indoleamine 2,3-dioxygenase (IDO), and (d) FoxP3 in rectal mucosa from vaccinated (n=6) and mock-vaccinated (n=5) animals. Bars represent the average for each group±standard deviation. All values in figure (a)–(d) are normalized on the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. (e) Correlative analysis between logarithmic values for FoxP3 and CTLA4 mRNA in the rectal mucosa of vaccinated or (f) mock-vaccinated animals at weeks 12, 16, and 24 post-infection. (g) Kyn/Trp ratio measured by high-performance liquid chromatography (HPLC) in the plasma of vaccinated, and (h) mock-vaccinated animals at weeks 7, 12, and 16 and 24 post-infection. (i) Mean values of Kyn/Trp plasma ration in all animals overtime.
To assess IDO activity more directly, we measured by high-performance liquid chromatography the plasma levels of kynurenine and tryptophan (Kyn/Trp) and calculated their ratio. Vaccination was associated with a significant lower plasma level of kynurenine/tryptophan ratio compared to the controls. This difference was already observed at week 7 and was maintained trough week 24 after infection, despite similar level of IDO expression in tissues at weeks 12 and 24 (Figure 5c and g–i). Considered together, these data suggest that the faster and higher level of expression of immune modulators in control macaques may be associated to increased virus level particularly at mucosal sites.
Discussion
Mucosal compartments encounter a plethora of antigenic stimuli, including pathogens. The appropriate activation of innate or adaptive effector mechanisms determines the ability of the host to fence off pathogens while avoiding bystander tissue damage and immunological exhaustion. A large pool of activated terminally differentiated memory effector CCR5+ CD4+ T cells reside in the lamina propria of mucosal tissues. HIV and SIV preferentially infect these memory CD4 T cells in mucosal effector sites, causing a rapid loss of this population within the first few weeks of infection.1, 2, 3 and 4 Over time, the CD4+ T-cell loss in SIV-infected macaques is associated with opportunistic infection and alteration in the intestinal structure and function.38 Thus, vaccines for HIV should likely elicit memory CD8+ T-cell responses that home at mucosal sites and are of sufficient magnitude to prevent or a least temper the loss of the CD4+ T cells in this location.
In nonhuman primate models of HIV, systemic vaccination regimens based on recombinant poxviruses17, 18, 19, 20, 27 and 31 or other live virus, vectors such as adenovirus21, 28 and 29 induce measurable CD8+ T-cell responses in the gut-associated lymphoid tissues and reduce viral replication. An immunization regimen combining DNA- and recombinant adenovirus-based vaccines has been tested for its efficacy in protecting from the loss of CD4 T help in jejunum.28, 29 In that study, the frequency of CD4+ and CD8+ T cells was derived from isolated mono-nuclear cells obtained from minced tissues, and the conclusion was reached that vaccination conferred protection from CD4+ T-cell loss.
Here we have evaluated the ability of vaccination with a DNA prime–MVA boost regimen to protect mucosal site from CD4+ T-cell loss and analyzed not only mononuclear cell suspension from minced tissues, but also in intact tissues by immune histochemistry. We found that CD4+ T cells were not spared in the vaccinated group despite the ability of this vaccine regimen to induce mucosal immune responses that were expanded following SIVmac251 infection and to reduce of 1,000-fold virus levels at mucosal site. In fact, a loss of up to 70% of CD4+ T cells was observed in both vaccinated and control macaques (Figure 4b, d and e). In our study, vaccination with DNA-MVA has reduced the virus to a similar, if not greater extent, to that reported in the case of the DNA-Ad5 vaccine platform,28, 29 ruling out that the difference reported here is to a lower level of protection induced by the vaccines used in our study.
The lack of protection from CD4+ T-cell loss afforded by vaccination is not unexpected as only antibodies that block infection could protect the large number of activated CCR5+ target T cells present at the site of exposure. Rather, the data suggest that most likely the number of the founder viruses is not affected by vaccination. This is in line with the hypothesis that the adaptive CD8+ T-cell responses may expand too late to curtail the founder virus.39 At later time points however, the expanded CD8+ T-cell responses may limit the local and systemic dissemination of SIV. Appropriate studies in vaccinated animals need to be designed to assess in detail the validity of this hypothesis.
We observed here that vaccination, by decreasing the overall viral burden at mucosal sites, favored the preservation of the CD4+ T cells that migrate to mucosa sites. This was observed only when CD3+ CD4+ CCR5+ cells were enumerated in mononuclear cell suspension. However, we found only a trend for a better preservation of CD4+ CCR5+ T cells in intact tissue. This discrepancy may be due to the fact that we were unable to co-stain CD4+ CCR5+ cells in tissues with antibody to CD3, a marker that readily identifies T cells.
Why are the CD4+ T cells that replenish the mucosal tissue better preserved in vaccinated animals? We could not demonstrate a clear difference in the activation of the host immune response by enumerating CD4+ Ki67+ T cells in mucosal tissues of vaccinated or mock-vaccinated macaques, suggesting that this activation marker “per se” does not inform on immune activation. Rather, we found differences in the expression of molecules that are associated with both immune activation and immune suppression, such as CTLA-4, IDO, FoxP3, and TGF-β, following challenge exposure to SIVmac251. The activation of these immune suppressive mechanisms is directly associated to exposure to the virus and to the level of viral replication.34 Therefore, the ability of vaccination to limit viral replication may directly account for the lesser and delayed activation of these immune-regulatory mechanisms in the vaccinated macaques. The finding of a direct correlation in the level of CTLA-4 and FoxP3 in mock, but not vaccinated animals, suggests an important difference in the quality of local innate and perhaps adaptive responses. Thus, it is possible that vaccine-induced responses, by limiting viral replication, may limit the induction of immune suppressive mechanisms at the mucosal sites, which in turn further favors the maintenance of an efficient antiviral response that contains viral replication.
Methods
Animals, treatments, and SIVmac251 challenge. All animals used in this study were colony-bred rhesus macaques (Macaca mulatta), obtained from Covance Research Products (Alice, TX). The animals were housed and handled in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International. The care and use of the animals were in compliance with all relevant institutional (NIH) guidelines.
A total of 13 macaques were enrolled and divided in two groups, as indicated in (Figure 1). Macaques in the first group (VACC) were immunized at weeks 0 and 4 with intramascular (3 mg) and intradermal (1 mg) inoculations of DNA-SIV-retanef and individuals plasmid DNAs encoding the codon-optimized SIV gag, pol, and envelope genes all given at different sites. For the boost, two poxvirus recombinants were used: vT360, a recombinant MVA that expresses SIVmac251 env, gag-pol genes and vT360, a recombinant MVA that expresses SIVmac239 retanef genes30 (Therion Biologics). All the seven animals were then boosted at weeks 26 and 52 with MVA-SIV-retanef and MVA-SIV-gpe. Each vaccination with MVA consisted of 108 PFU in a volume of 150 μl of phosphate-buffered saline delivered intramuscularly at two sites for each animal. Animals in the second group (MOCK) were mock vaccinated with the equivalent amount of nonrecombinant MVA.
Preparation of lymphocytes from blood and tissues. Mononuclear cells from blood and LNs were isolated by density-gradient centrifugation on Ficoll and resuspended in RPMI 1640 medium (Gibco BRL, Gaithersburg, MD) containing 10% fetal bovine serum (R-10). Rectal and vaginal pinches were treated with 1 mM of Ultra Pure Dithiothreitol (Invitrogen, Carlsbad, CA) for 30 min followed by incubation in calcium/magnesium-free Hank's balanced salt solution (Gibco BRL). Following the removal of epithelium and intraepithelial lymphocytes, the tissues were incubated with collagenase D (400 U/ml; Boehringer Mannheim, Mannheim, Germany) and DNase (1 μg/ml; Invitrogen) for 2.5 h at 37 °C in Iscove's modified Dulbecco's medium (Gibco BRL) supplemented with 10% fetal bovine serum and penicillin–streptomycin. The suspensions were shacked every 15 min during this incubation period. The dissociated mononuclear cells were than placed over 42% Percoll (General Electric Healthcare, Piscataway, NJ) and centrifuged at 800g for 25 min at 4 °C. Lamina propria lymphocytes were collected from the cell pellet.
Viral RNA in plasma and tissues. SIVmac251 in plasma was quantified by nucleic acid sequence-based amplification.40 Briefly, RNA was extracted from plasma and tissues as previously described41 and isothermally amplified using SIVmac251-specific primers. Quantification was conducted by using an electrochemiluminescence chemistry-based probe hybridization system with a coextracted internal standard. The copy number was expressed per 100 μl of plasma or per microgram of RNA, and the detection limit of the assay was 2 × 103 RNA copies.
Total RNA from LNs mononuclear cells and from rectal pinches biopsies was extracted using the Trizol LS Reagent (Invitrogen), according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed into first-strand cDNA in a 20 μl reaction containing 1 μM random hexanucleotide primers, 1 μM oligo dT, and 200 U Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI).
cDNA quantification for IDO, TGF-β, CTLA-4, FOXP3, SIV Gag RNA, and glyceraldehyde 3-phosphate dehydrogenase was performed by real-time PCR conducted with the ABI Prism 7900HT (Applied Biosystems, Foster City, CA). All reactions were performed using a SYBR Green PCR mix (Qiagen, Valencia, CA), as previously described (CTLA4). Data analysis was performed with the SDS2.1 software, provided with the ABI Prism 7900HT. Primer sequences were designed using the Primer3 software as previously described.35
Immune histochemistry and immune fluorescence. Tissue was fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and embedded in paraffin. All slides were stained using the Dako Autostainer (Dako, Carpenteria, CA). Tris-buffered saline with Tween 20 (Dako) was used for all washes. Bovine serum albumin/monkey serum/Tris-buffered saline (5%) was used for all antibody dilutions. The primary antibodies used included monoclonal antibody (mAb) anti-CD4 mouse IgG1 (clone IF6; Vector, Burlingame, CA), biotinylated mAb anti-CD45RA mouse IgG1 (clone F8-11-13; AbD Serotec, Raleigh, NC), polyclonal anti-CD95 rabbit serum (Genetex, San Antonio, TX), and polyclonal anti-CCR5 rabbit serum (ProSci, Poway, CA). For all primary antibodies, slides were subjected to an antigen retrieval step consisting of incubation in citrate (pH 6.0; Dako) for 2 min at 124 °C in the Digital Decloaking Chamber (Biocare Medical, Concord, CA) followed by cooling for 20 min to 89 °C before rinsing in running water and a final buffer rinse. Primary antibodies were replaced by normal rabbit immunoglobulin G (IgG; Zymed, South San Francisco, CA) or mouse IgG (Dako) and included with each staining series as the negative control. For double-label immunofluorescence, nonspecific binding sites were blocked with 10% goat serum and Tween 20 in phosphate-buffered saline (Background Eraser; Biocare Medical). Binding of the primary antibodies was detected simultaneously using Alexa Fluor 488-labeled polyclonal goat anti-rabbit IgG (Molecular Probes, Eugene OR), Alexa Fluor 568-labeled polyclonal goat anti-mouse IgG (Molecular Probes), or streptavidin Alexa Fluor 568 conjugate for 1 h (Molecular Probes). Nonspecific binding sites were further blocked by the addition of a 30 min incubation in 5% bovine serum albumin/Tris-buffered saline between the first primary antibody incubation and detection reagents, and the addition of second primary antibody and detection reagents. All slides were coverslipped using ProLong Gold with 4′,6-diamidino-2-phenylindole dihydrochloride hydrate (Molecular Probes, Carlsbad, CA) to stain nuclei. All the control experiments gave appropriate results with minimal nonspecific staining (data not shown).
Slides were visualized with epi-fluorescent illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss Inc., Thornwood, NY) and appropriate filters. Digital images were captured and analyzed by using a Zeiss Axiocam System and Openlab software (Inprovision, Waltham, MA).
Quantitative image analysis. The number of positive mAb-labeled cells/mm2 of tissue sample was quantified using a Zeiss AxioCam HRc digital camera mounted on a Zeiss microscope (Zeiss, Jena, Germany) fitted with a × 40 plan neofluar objective and a polarizing filter cube with appropriate filters (Omega Optical, Brattleboro, VT). Digital images were captured with Openlab software (Improvision, Lexington, MA). One section per tissue with representative histomorphological components (cortex, paracortex, follicles, medulla for LN; and epithelium, lamina propria, and muscularis mucosa for rectum) was analyzed. Five high-power (× 40) microscope fields of the T-cell-rich zone (paracortex) per LN section and five high-power fields of lamina propria from rectal sections were randomly chosen and captured digitally with the system described above. Each captured field includes an area of approximately 0.04 mm2. Only clearly positive cells with distinctly labeled nuclei (4′,6-diamidino-2-phenylindole dihydrochloride hydrate) and bright staining were considered positive. Individual positive cells in the five captured high-power microscope fields of the immunohistochemical stained tissue sections were counted manually by a single observer. The numbers of positive cells are presented as cells per square millimeter.
Tryptophan and kynurenine concentration measurement by high-performance liquid chromatography. Detection of tryptophan and kynurenine was performed on plasma collected from animals by high-performance liquid chromatography as previously described.34, 42
Statistical analysis. Statistical analyses for mRNA expression, tryptophan and kynurenine ratio, and LN viral load were performed using the SPSS 13.0 software (SPSS, Chicago, IL). Differences between groups were assessed by nonparametric Mann–Whitney U-test. Differences before and after treatment within the same group were assessed using the Wilcoxon test. All P-values shown in the text and figures are two tailed.
CD4+ and CD8+ T-cell counts. CD4+ and CD8+ counts were periodically determined whole blood and by fluorescence-activated cell sorting analysis, according to the FACS/Lyse kit (BD Immunocytometry Systems, San Jose, CA), and analysis were performed using a FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA). The antibodies used were CD3 (BD Biosciences; SP34), CD4 (BD Biosciences; L200), CD8 (DK25; DakoCytomation, Carpinteria, CA), and CD20 (B9E9; Beckman Coulter, Fullerton, CA).
Lymphocyte proliferation assay. Peripheral blood mononuclear cell were cultured at 105 cells per well in triplicates for 3 days in the absence or presence of native high-performance liquid chromatography-purified SIV p27 Gag or gp120 Env proteins (Advanced BioScience Laboratories, Kensington, MD), or concavalin-A as a positive control. The cells were then pulsed overnight with 1 μCi of [3H] thymidine before harvest. The relative rate of lymphoproliferation was calculated as fold of thymidine incorporation into cellular DNA over medium control (stimulation index).
Intracellular staining and ELISPOT. SIV-specific CD4+ and CD8+ T-cell responses were detected using pools of 15-meric peptides overlapping by 11 amino acids covering entire Gag, Env, and Pol proteins of SIVmac239. Cells (1 × 106) in RPMI 1640 medium (containing 10% human serum and antibiotics) were incubated in the absence or the presence of a specific peptide pool at 1 μg/ml of each peptide for 1 h or in the presence of the super-antigen staphylococcal enterotoxin B (Sigma, St. Louis, MO, US) at a 1 mg/ml final concentration as a positive control. The co-stimulatory mAb CD28 and CD49d (0.5 μg/ml; BD Pharmingen, San Diego, CA) were added to all the samples to maximize the detection of T cells with higher activation thresholds. Positive control was treated with staphylococcal enterotoxin B Brefeldin A (GolgiPlug; BD Biosciences) at a final concentration of 10 μg/ml was added, and the cells were incubated for an additional 5 h. The cells were washed, stained for the surface Ags with anti-human CD3ɛ (clone SP34; BD Pharmingen) and anti-human CD8β Abs (clone 2ST8.5H7; Beckman Coulter), permeabilized by incubation in Cytofix/Cytoperm solution (BD Biosciences), and stained with anti-human IL2-FITC (clone MQ1-17H12), and anti-IFN-γ-APC mAbs (clone B27; BD Pharmingen). The results were calculated as the total number of cytokine-positive cells with background subtracted.
ELISPOT was performed using the macaque IFN-γ-specific ELISPOT kits (U-Cytech, Utrecht, the Netherlands) were used to detect the number of Gag-specific, IFN-γ-producing cells. Ninety-six-well, flat-bottom plates were coated with anti-IFN-γ mAb MD-1 overnight at 4 °C and blocked with 2% bovine serum albumin in phosphate-buffered saline for 1–3 h at 37 °C. Cells (1 × 105/well) were loaded in triplicate in RPMI 1640 containing 5% human serum and specific peptides or 5 μg/ml concavalin-A as a positive control. The plates were incubated overnight at 37 °C in 5% CO2 and developed according to the manufacturer's guidelines (U-Cytech).
Flow cytometry. For the physical detection of SIV-specific CD8+ T cells, staining was performed with anti-human CD8β-PE, anti-human CD28-FITC (clone CD28.2; BD Pharmingen), anti-human CD95-PECy5 mAbs (clone DX2; BD Pharmingen), and with pretitered, allophycocyanin (APC)-conjugated (Molecular Probes) Gag181–189 CM9 (p11C) (CTPYDINQM)–Mamu-A*01 tetrameric complexes (Beckman Coulter) for 30 min at room temperature.43 A total of 100,000 events were collected in the lymphocyte region (R1) and analyzed with CellQuest software (BD Biosciences). For phenotypic analysis of the CD4 T-cell subsets, cells were labeled simultaneously with the following combinations of anti-human antibodies: CD3ɛ-FITC, CD4-PerCP (clone L200; BD Biosciences), CD195 (CCR5)-PE (3A9; BD Pharmingen), CD95-PECy5 and CD45RA-FITC Abs (clone HI100; BD Pharmingen). All antibodies were validated and titrated using rhesus macaque peripheral blood mononuclear cells. For the intracellular staining, cells were labeled as described, fixed with 0.5% paraformaldehyde, and analyzed using a FACSCalibur from Becton Dickinson. A total of 50,000 events in the lymphocyte region were collected. Data were analyzed using CellQuest (Beckton Dickinson) or FlowJo version 6.1 (Tree Star, Ashland, OR, USA).
Disclosure
The authors declared no conflict of interest.
References
Veazey, R.S. et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280, 427–431 (1998).
Li, Q. et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434, 1148–1152 (2005).
Mattapallil, J.J. et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434, 1093–1097 (2005).
Veazey, R.S. et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4(+) T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74, 57–64 (2000).
Grivel, J.C. & Margolis, L.B. CCR5 and CXCR4-tropic HIV are equally cytopatic for their T-cell targets in human lymphoid tissues. Nat. Med. 5, 344–346 (1999).
Lee, B., Sharron, M., Montaner, L.J., Weissman, D. & Doms, R.W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. 96, 5215–5220 (1999).
Marx, P.A. & Chen, Z. The function of simian chemokine receptor in the replication of SIV. Semin. Immunol. 10, 215 (1998).
Mehandru, S. et al. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J. Virol. 81, 599–612 (2007).
Picker, L.J. et al. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200, 1299–1314 (2004).
Guadalupe, M. et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77, 11708–11717 (2003).
Guadalupe, M. et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J. Virol. 80, 8236–8247 (2006).
Paiardini, M. et al. Perturbations of cell cycle control in T cells contribute to the different outcomes of simian immunodeficiency virus infection in rhesus macaques and sooty mangabeys. J. Virol. 80, 10335–10345 (2006).
Pandrea, I. et al. Acute loss of intestinal CD4 T cells is not predictive of simian immunodeficiency virus virulence. J. Immunol. 179, 3035–3046 (2007).
Grossman, Z. et al. The impact of HIV on naïve T-cell homeostasis. Nat. Med. 6, 976–977 (2000).
Ho, D. et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 117–122 (1995).
Sousa, A. et al. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 169, 3400–3406 (2002).
Hel, Z. et al. Potentiation of simian immunodeficiency virus (SIV)-specific CD4(+) and CD8(+) T cell responses by a DNA-SIV and NYVAC-SIV prime/boost regimen. J. Immunol. 167, 7180–7191 (2001).
Hel, Z. et al. Containment of simian immunodeficiency virus infection in vaccinated macaques: correlation with the magnitude of virus-specific pre- and postchallenge CD4+ and CD8+ T cell responses. J. Immunol. 169, 4778–4787 (2002).
Seth, A. et al. Recombinant modified vaccinia virus Ankara-simian immunodeficiency virus gag pol elicits cytotoxic T lymphocytes in rhesus monkeys detected by a major histocompatibility complex class I/peptide. Proc. Natl. Acad. Sci. 95, 10112–10116 (1998).
Amara, R.R. et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292, 69–74 (2001).
Baig, J. et al. Elicitation of simian immunodeficiency virus-specific cytotoxic T lymphocytes in mucosal compartments of rhesus monkeys by systemic vaccination. J. Virol. 76, 11484–11490 (2002).
Stevceva, L. et al. Both mucosal and systemic routes of immunization with the live, attenuated NYVAC/simian immunodeficiency virus SIV(gpe) recombinant vaccine result in gag specific CD8(+) T cell responses in mucosal tissues of macaques. J. Virol. 76, 11659–11676 (2002).
Wherry, E.J. et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4, 225–234 (2002).
Matloubian, M. et al. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68, 8056–8065 (1994).
Shedlock, D.J. & Shen, H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300, 337–339 (2003).
Toellner, K.S. et al. Th1 and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J. Exp. Med. 187, 1193–12011 (1998).
Vaccari, M. et al. Reduced protection from SIV afforded by memory CD8+T-cells induced by vaccination in condition of CD4+T-cell deficiency. J. Virol. (in press).
Mattapalli, J.J. et al. Systemic vaccination prevents the total destruction of mucosal CD4 T cells during acute SIV challenge. J. Med. Primatol. 35, 217–224 (2006).
Mattapallil, J.J. et al. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J. Exp. Med. 203, 1533–1541 (2006).
Hel, Z. et al. A novel chimeric Rev, Tat, and Nef (Retanef) antigen as a component of an SIV/HIV vaccine. Vaccine 20, 3171–3186 (2002).
Hel, Z. et al. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J. Immunol. 176, 85–96 (2006).
Pitcher, J. et al. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168, 29–43 (2002).
Vaccari, M. et al. Vaccine-induced CD8+ central memory T-cells in protection from simian AIDS. J. Immunol. 175, 3502–3507 (2005).
Boasso, A. et al. Regulatory T-cell markers, indoleamine 2,3-dioxygenase, and virus levels in spleen and gut during progressive simian immunodeficiency virus infection. J. Virol. 81, 11593–11603 (2007).
Hryniewicz, A. et al. CTLA-4 blockade decreases TGF-beta, IDO, and viral RNA expression in tissues of SIVmac251-infected macaques. Blood 108, 3834–3842 (2006).
Hartigan-O'Connor, D.J., Abel, K. & McCune, J.M. Suppression of SIV-specific CD4+ T cells by infant but not adult macaque regulatory T cells: implications for SIV disease progression. J. Exp. Med. 204, 2679–2692 (2007).
Tang, Q., Boden, E.K., Henriksen, K.J., Bour-Jordan, H., Bi, M. & Bluestone, J.A. Distinct roles of CTLA-4 and TGF-beta in CD4+CD25+ regulatory T cell function. Eur. J. Immunol. 34, 2996–3005 (2004).
Heise, C., Miller, C.J., Lackner, A. & Dandekar, S. Primary acute simian immunodeficiency virus infection of intestinal lymphoid tissue is associated with gastrointestinal dysfunction. J. Infect. Dis. 169, 1116–1120 (1994).
Reynolds, M.R. et al. CD8+ T-lymphocyte response to major immunodominant epitopes after vaginal exposure to simian immunodeficiency virus: too late and too little. J. Virol. 79, 9228–9235 (2005).
Romano, J.W., Williams, K.G., Shurtliff, R.N., Ginocchio, C. & Kaplan, M. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 26, 15–28 (1997).
Pal, R. et al. Systemic immunization with an ALVAC-HIV-1/protein boost vaccine strategy protects rhesus macaques from CD4+ T-cell loss and reduces both systemic and mucosal simian-human immunodeficiency virus SHIVKU2 RNA levels. J. Virol. 80, 3732–3742 (2006).
Widner, B., Werner, E.R., Shennach, H., Wachter, H. & Fuchs, D. Simultaneous measurement of serum tryptophan and kynurenine by HPLC. Clin. Chem. 43, 2424–2426 (1997).
Kuroda, M.J. et al. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J. Exp. Med. 187, 1373–1381 (1999).
Acknowledgements
We thank P.D. Markham, S. Orndorff, D. Weiss, and J. Treece of Advanced BioScience Laboratories, Kensington, MD. We thank D. Panicali from Therion for constructing and providing us the recombinant poxviruses. We also thank Steven Snodgrass, Ryan Kelly, and Teresa Habina for the editorial assistance. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vaccari, M., Boasso, A., Ma, ZM. et al. CD4+ T-cell loss and delayed expression of modulators of immune responses at mucosal sites of vaccinated macaques following SIVmac251 infection. Mucosal Immunol 1, 497–507 (2008). https://doi.org/10.1038/mi.2008.60
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2008.60
This article is cited by
-
Comparison of systemic and mucosal vaccination: impact on intravenous and rectal SIV challenge
Mucosal Immunology (2012)
-
Use of Nonhuman Primate Models to Develop Mucosal AIDS Vaccines
Current HIV/AIDS Reports (2010)