Abstract
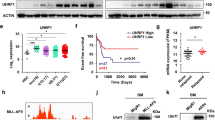
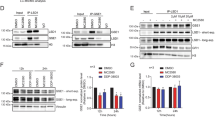
Lysine-specific demethylase 1 (LSD1) regulates gene expression by affecting histone modifications and is a promising target for acute myeloid leukemia (AML) with specific genetic abnormalities. Novel LSD1 inhibitors, NCD25 and NCD38, inhibited growth of MLL-AF9 leukemia as well as erythroleukemia, megakaryoblastic leukemia and myelodysplastic syndromes (MDSs) overt leukemia cells in the concentration range that normal hematopoiesis was spared. NCD25 and NCD38 invoked the myeloid development programs, hindered the MDS and AML oncogenic programs, and commonly upregulated 62 genes in several leukemia cells. NCD38 elevated H3K27ac level on enhancers of these LSD1 signature genes and newly activated ~500 super-enhancers. Upregulated genes with super-enhancer activation in erythroleukemia cells were enriched in leukocyte differentiation. Eleven genes including GFI1 and ERG, but not CEBPA, were identified as the LSD1 signature with super-enhancer activation. Super-enhancers of these genes were activated prior to induction of the transcripts and myeloid differentiation. Depletion of GFI1 attenuated myeloid differentiation by NCD38. Finally, a single administration of NCD38 causes the in vivo eradication of primary MDS-related leukemia cells with a complex karyotype. Together, NCD38 derepresses super-enhancers of hematopoietic regulators that are silenced abnormally by LSD1, attenuates leukemogenic programs and consequently exerts anti-leukemic effect against MDS-related leukemia with adverse outcome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Shih AH, Abdel-Wahab O, Patel JP, Levine RL . The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012; 12: 599–612.
Jenuwein T, Allis CD . Translating the histone code. Science 2001; 293: 1074–1080.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39: 311–318.
He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet 2010; 42: 343–347.
Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 2013; 51: 310–325.
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proce Natl Acad Sci USA 2010; 107: 21931–21936.
Voigt P, Tee WW, Reinberg D . A double take on bivalent promoters. Genes Dev 2013; 27: 1318–1338.
Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159: 1312–1326.
Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014; 159: 1327–1340.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319.
Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 2014; 345: 943–949.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C et al. A novel mammalian flavin-dependent histone demethylase. J Biol Chem 2009; 284: 17775–17782.
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y . Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005; 19: 857–864.
Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O'Brien K et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. ELife 2013; 2: e00633.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947.
Niebel D, Kirfel J, Janzen V, Holler T, Majores M, Gutgemann I . Lysine-specific demethylase 1 (LSD1) in hematopoietic and lymphoid neoplasms. Blood 2014; 124: 151–152.
Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 2012; 18: 605–611.
Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012; 21: 473–487.
Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 2014; 28: 2155–2164.
Gelato KA, Shaikhibrahim Z, Ocker M, Haendler B . Targeting epigenetic regulators for cancer therapy: modulation of bromodomain proteins, methyltransferases, demethylases, and microRNAs. Expert Opin Ther Targets 2016; 20: 783–799.
Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012; 120: 2454–2465.
Santos FP, Faderl S, Garcia-Manero G, Koller C, Beran M, O'Brien S et al. Adult acute erythroleukemia: an analysis of 91 patients treated at a single institution. Leukemia 2009; 23: 2275–2280.
Oki Y, Kantarjian HM, Zhou X, Cortes J, Faderl S, Verstovsek S et al. Adult acute megakaryocytic leukemia: an analysis of 37 patients treated at M.D. Anderson Cancer Center. Blood 2006; 107: 880–884.
Ogasawara D, Itoh Y, Tsumoto H, Kakizawa T, Mino K, Fukuhara K et al. Lysine-specific demethylase 1-selective inactivators: protein-targeted drug delivery mechanism. Angew Chem Int Ed 2013; 52: 8620–8624.
Tohyama K, Tohyama Y, Nakayama T, Ueda T, Nakamura T, Yoshida Y . A novel factor-dependent human myelodysplastic cell line, MDS92, contains haemopoietic cells of several lineages. Br J Haematol 1995; 91: 795–799.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137.
Anders S, Huber W . Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013; 14: 128.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44: W90–W97.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823.
Brown AL, Wilkinson CR, Waterman SR, Kok CH, Salerno DG, Diakiw SM et al. Genetic regulators of myelopoiesis and leukemic signaling identified by gene profiling and linear modeling. J Leukocyte Biol 2006; 80: 433–447.
Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 2009; 113: 2375–2385.
Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 2014; 25: 652–665.
Kuo HP, Wang Z, Lee DF, Iwasaki M, Duque-Afonso J, Wong SH et al. Epigenetic roles of MLL oncoproteins are dependent on NF-kappaB. Cancer Cell 2013; 24: 423–437.
Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol 2008; 9: 810–819.
Reid S, Ritchie A, Boring L, Gosling J, Cooper S, Hangoc G et al. Enhanced myeloid progenitor cell cycling and apoptosis in mice lacking the chemokine receptor, CCR2. Blood 1999; 93: 1524–1533.
Iida S, Watanabe-Fukunaga R, Nagata S, Fukunaga R . Essential role of C/EBPalpha in G-CSF-induced transcriptional activation and chromatin modification of myeloid-specific genes. Genes Cells 2008; 13: 313–327.
Richard M, Veilleux P, Rouleau M, Paquin R, Beaulieu AD . The expression pattern of the ITIM-bearing lectin CLECSF6 in neutrophils suggests a key role in the control of inflammation. J Leukocyte Biol 2002; 71: 871–880.
Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 2002; 30: 295–300.
Moignard V, Macaulay IC, Swiers G, Buettner F, Schutte J, Calero-Nieto FJ et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol 2013; 15: 363–372.
Guo G, Luc S, Marco E, Lin TW, Peng C, Kerenyi MA et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 2013; 13: 492–505.
Saleque S, Kim J, Rooke HM, Orkin SH . Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol cell 2007; 27: 562–572.
Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 2003; 18: 109–120.
Tenen DG . Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer 2003; 3: 89–101.
Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014; 157: 369–381.
Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 2014; 25: 415–427.
Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci USA 2009; 106: 10141–10146.
Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 2013; 153: 1149–1163.
Acknowledgements
We are grateful to Dr Toshio Kitamura (University of Tokyo) for kindly gifting the vectors and providing scientific comments and to Dr Kaoru Tohyama (Kawasaki Medical School) and Dr Akihiko Yokoyama (Kyoto University) for kindly gifting the cell lines and providing scientific comments. This work was supported by the JSPS KAKENHI grant numbers JP25118713 and JP16K09846 (MK), the Kanae Foundation for the promotion of medical science (MK), and the Takeda Science Foundation (MK). This work was carried out at the Joint Usage/Research Center (RIRBM), Hiroshima University. This work was funded by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MK), the Kanae Foundation for the promotion of medical science (MK), and the Takeda Science Foundation (MK).
Author contributions
NS performed and analyzed most of experiments and wrote the manuscript. MK obtained grants, organized the project, designed and analyzed all experiments, and wrote the manuscript. GT performed and analyzed the ChIP experiments and GFI1-knockout experiments. AK, HM and TI performed and analyzed the ChIP-seq. RY performed WB. YN, SF, YS and MH assisted with some experiments. TS provided the compounds. AA and AT supervised the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Sugino, N., Kawahara, M., Tatsumi, G. et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia 31, 2303–2314 (2017). https://doi.org/10.1038/leu.2017.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.59
This article is cited by
-
Role of HOXA9 in solid tumors: mechanistic insights and therapeutic potential
Cancer Cell International (2022)
-
Combining LSD1 and JAK-STAT inhibition targets Down syndrome-associated myeloid leukemia at its core
Leukemia (2022)
-
KDM1A inhibition is effective in reducing stemness and treating triple negative breast cancer
Breast Cancer Research and Treatment (2021)
-
LSD1-mediated repression of GFI1 super-enhancer plays an essential role in erythroleukemia
Leukemia (2020)
-
Merkel cell polyomavirus activates LSD1-mediated blockade of non-canonical BAF to regulate transformation and tumorigenesis
Nature Cell Biology (2020)