Abstract
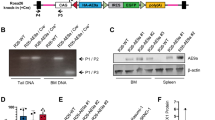
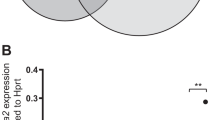
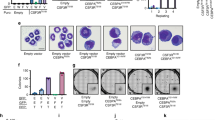
Acute megakaryoblastic leukemia in patients without Down syndrome is a rare malignancy with a poor prognosis. RNA sequencing of fourteen pediatric cases previously identified novel fusion transcripts that are predicted to be pathological including CBFA2T3-GLIS2, GATA2-HOXA9, MN1-FLI and NIPBL-HOXB9. In contrast to CBFA2T3-GLIS2, which is insufficient to induce leukemia, we demonstrate that the introduction of GATA2-HOXA9, MN1-FLI1 or NIPBL-HOXB9 into murine bone marrow induces overt disease in syngeneic transplant models. With the exception of MN1, full penetrance was not achieved through the introduction of fusion partner genes alone, suggesting that the chimeric transcripts possess a unique gain-of-function phenotype. Leukemias were found to exhibit elements of the megakaryocyte erythroid progenitor gene expression program, as well as unique leukemia-specific signatures that contribute to transformation. Comprehensive genomic analyses of resultant murine tumors revealed few cooperating mutations confirming the strength of the fusion genes and their role as pathological drivers. These models are critical for both the understanding of the biology of disease as well as providing a tool for the identification of effective therapeutic agents in preclinical studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Creutzig U, Reinhardt D, Diekamp S, Dworzak M, Stary J, Zimmermann M . AML patients with Down syndrome have a high cure rate with AML-BFM therapy with reduced dose intensity. Leukemia 2005; 19: 1355–1360.
Athale UH, Razzouk BI, Raimondi SC, Tong X, Behm FG, Head DR et al. Biology and outcome of childhood acute megakaryoblastic leukemia: a single institution's experience. Blood 2001; 97: 3727–3732.
Barnard DR, Alonzo TA, Gerbing RB, Lange B, Woods WG . Comparison of childhood myelodysplastic syndrome, AML FAB M6 or M7, CCG 2891: report from the Children's Oncology Group. Pediatr Blood Cancer 2007; 49: 17–22.
Garderet L, Labopin M, Gorin NC, Polge E, Baruchel A, Meloni G et al. Hematopoietic stem cell transplantation for de novo acute megakaryocytic leukemia in first complete remission: a retrospective study of the European Group for Blood and Marrow Transplantation (EBMT). Blood 2005; 105: 405–409.
Reinhardt D, Diekamp S, Langebrake C, Ritter J, Stary J, Dworzak M et al. Acute megakaryoblastic leukemia in children and adolescents, excluding Down's syndrome: improved outcome with intensified induction treatment. Leukemia 2005; 19: 1495–1496.
O'Brien MM, Cao X, Pounds S, Dahl GV, Raimondi SC, Lacayo NJ et al. Prognostic features in acute megakaryoblastic leukemia in children without Down syndrome: a report from the AML02 multicenter trial and the Children's Oncology Group Study POG 9421. Leukemia 2013; 27: 731–734.
Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 2002; 32: 148–152.
Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN . RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 2003; 101: 4333–4341.
Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet 2013; 45: 1293–1299.
Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, Ta HQ et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012; 22: 683–697.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP . Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012; 7: e46688.
Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res 1995; 23: 628–633.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764.
Olshen AB, Venkatraman ES, Lucito R, Wigler M . Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004; 5: 557–572.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Gruber TA, Downing JR. Molecular genetics of acute myeloid leukemia. In: Pui CH (ed). Childhood Leukemias, 3rd edn. Cambridge University Press: New York, USA, 2012, pp 204–238..
Westervelt P, Lane AA, Pollock JL, Oldfather K, Holt MS, Zimonjic DB et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood 2003; 102: 1857–1865.
Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 1996; 85: 853–861.
Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell 2011; 20: 597–609.
Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ . EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol 2007; 27: 7918–7934.
Wang GG, Pasillas MP, Kamps MP . Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood 2005; 106: 254–264.
Huang Z, Dore LC, Li Z, Orkin SH, Feng G, Lin S et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol 2009; 29: 5168–5180.
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467: 430–435.
Kandilci A, Surtel J, Janke L, Neale G, Terranova S, Grosveld GC . Mapping of MN1 sequences necessary for myeloid transformation. PLoS One 2013; 8: e61706.
Heuser M, Argiropoulos B, Kuchenbauer F, Yung E, Piper J, Fung S et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood 2007; 110: 1639–1647.
Lai CK, Moon Y, Kuchenbauer F, Starzcynowski DT, Argiropoulos B, Yung E et al. Cell fate decisions in malignant hematopoiesis: leukemia phenotype is determined by distinct functional domains of the MN1 oncogene. PLoS One 2014; 9: e112671.
Braunholz D, Hullings M, Gil-Rodriguez MC, Fincher CT, Mallozzi MB, Loy E et al. Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction. Eur J Hum Genet 2012; 20: 271–276.
Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR . Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 2002; 1: 63–74.
Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J 2005; 24: 368–381.
Mercher T, Raffel GD, Moore SA, Cornejo MG, Baudry-Bluteau D, Cagnard N et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest 2009; 119: 852–864.
Salek-Ardakani S, Smooha G, de Boer J, Sebire NJ, Morrow M, Rainis L et al. ERG is a megakaryocytic oncogene. Cancer Res 2009; 69: 4665–4673.
Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M et al. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol 2003; 23: 3427–3441.
Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006; 442: 818–822.
Shah N, Sukumar S . The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10: 361–371.
de Rooij JDE, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017; e-pub ahead of print 23 January 2017 doi:10.1038/ng.3772.
Stirewalt DL, Radich JP . The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer 2003; 3: 650–665.
Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D . RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11: 761–774.
Wartman LD, Larson DE, Xiang Z, Ding L, Chen K, Lin L et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest 2011; 121: 1445–1455.
Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002; 30: 41–47.
Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 1996; 10: 1911–1918.
Volpe G, Walton DS, Del Pozzo W, Garcia P, Dasse E, O'Neill LP et al. C/EBPalpha and MYB regulate FLT3 expression in AML. Leukemia 2013; 27: 1487–1496.
Wang GG, Pasillas MP, Kamps MP . Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol 2006; 26: 3902–3916.
Hanzelmann S, Castelo R, Guinney J . GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14: 7.
Acknowledgements
We would like to thank the staff of Flow Cytometry Core, Veterinary Pathology, as well as the Hartwell Center for Biotechnology and Bioinformatics of St Jude Children’s Research Hospital. Jinghui Zhang and the department of computational biology graciously provided previously established bioinformatic pipelines and support. We thank Charles J Sherr, Thomas A Look and Jeffery M Klco for their comments on the manuscript. This work was supported by grants from the Eric Trump Foundation, Gabrielle Angel Foundation and the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Dang, J., Nance, S., Ma, J. et al. AMKL chimeric transcription factors are potent inducers of leukemia. Leukemia 31, 2228–2234 (2017). https://doi.org/10.1038/leu.2017.51
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.51
This article is cited by
-
c-Mpl-del, a c-Mpl alternative splicing isoform, promotes AMKL progression and chemoresistance
Cell Death & Disease (2022)
-
Menin is necessary for long term maintenance of meningioma-1 driven leukemia
Leukemia (2021)
-
High WBP5 expression correlates with elevation of HOX genes levels and is associated with inferior survival in patients with acute myeloid leukaemia
Scientific Reports (2020)
-
Long-read sequencing unveils IGH-DUX4 translocation into the silenced IGH allele in B-cell acute lymphoblastic leukemia
Nature Communications (2019)
-
A high-throughput screen indicates gemcitabine and JAK inhibitors may be useful for treating pediatric AML
Nature Communications (2019)