Abstract
Precise classification of acute leukemia (AL) is crucial for adequate treatment. EuroFlow has previously designed an AL orientation tube (ALOT) to guide towards the relevant classification panel (T-cell acute lymphoblastic leukemia (T-ALL), B-cell precursor (BCP)-ALL and/or acute myeloid leukemia (AML)) and final diagnosis. Now we built a reference database with 656 typical AL samples (145 T-ALL, 377 BCP-ALL, 134 AML), processed and analyzed via standardized protocols. Using principal component analysis (PCA)-based plots and automated classification algorithms for direct comparison of single-cells from individual patients against the database, another 783 cases were subsequently evaluated. Depending on the database-guided results, patients were categorized as: (i) typical T, B or Myeloid without or; (ii) with a transitional component to another lineage; (iii) atypical; or (iv) mixed-lineage. Using this automated algorithm, in 781/783 cases (99.7%) the right panel was selected, and data comparable to the final WHO-diagnosis was already provided in >93% of cases (85% T-ALL, 97% BCP-ALL, 95% AML and 87% mixed-phenotype AL patients), even without data on the full-characterization panels. Our results show that database-guided analysis facilitates standardized interpretation of ALOT results and allows accurate selection of the relevant classification panels, hence providing a solid basis for designing future WHO AL classifications.
Similar content being viewed by others
Introduction
Acute leukemias (AL) are malignant expansions of aberrant haematopoietic precursor cells arrested at very immature stages of differentiation. Based on cytomorphologic, cytochemical and flowcytometric investigations, two main subgroups of AL, that is, acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) can be distinguished. Further subdivision of ALL fully relies on immunophenotyping and allows identification of T-cell acute lymphoblastic leukemia (T-ALL) and B-cell precursor (BCP)-ALL.1, 2 Appropriate diagnosis of these three main AL types is clinically essential, since these groups (particularly ALL versus AML) differ significantly in treatment and prognosis.
Flowcytometric immunophenotyping is a key diagnostic tool to identify the lineage of leukemic cells in AL patients. In a small number of cases, appropriate lineage assessment can be tricky, since the leukemic cells show no clear evidence of differentiation along a single lineage or they express differentiation antigens highly specific for more than one lineage.3 These cases account for <5% of all acute leukemia (AL) cases and they are categorized separately in the current WHO classification as AL of ambiguous lineage, including both acute undifferentiated leukemia (AUL) and mixed phenotype AL (MPAL).3, 4, 5 For MPAL cases, the WHO classification has defined the phenotypic criteria for cell lineage assignment, that is, CD19 and Cytoplasmic (Cy)CD79a, CD10 and/or CD22 for B-cells, CyCD3 or surface membrane (Sm)CD3 for T-cells, and myeloperoxidase (CyMPO) or monocytic differentiation markers (for example, CD14, CD64, CD11c) for myeloid lineage.3, 6
Within EuroFlow, we have developed fully standardized protocols and antibody panels for the diagnosis classification and monitoring of hematological malignancies.7, 8, 9, 10 For AL, a single eight-color tube (AL orientation tube, ALOT) was designed to orientate towards the complementary panels for full characterization of AL of different T, B and myeloid cell lineages. This tube contained the cell-lineage defining markers CyCD3, SmCD3, CD19, CyCD79a and CyMPO, as well as the pan-leukocyte marker CD45, the immature marker CD34 and CD7. Of note, CyCD79a was preferred over CyCD22 because CD22 is not lineage specific as it is also expressed at high levels in normal basophils, mast cells and some dendritic cells.11 CD7 was selected because it is positive in virtually all cases of T-ALL and in a subset of usually CyMPO-negative AML;7 in BCP-ALL CD7 is occasionally observed in a subset of MLL rearranged or Ph1pos BCP-ALLs.8 Despite ALOT is standardized in terms of both reagent composition, sample preparation, instrument set-up and data acquisition, final interpretation of the results still relies on local individual expertise and relatively subjective/arbitrary criteria for identification of leukemic cells and definition of positivity for individual markers.
Here we report on the design, construction and validation of a large ALOT database and the development of an automated database-guided analytical algorithm to support subsequent selection of the appropriate classification panel(s) for the diagnosis of AL patients. Standardized ALOT data7, 8 collected in the participating EuroFlow centers were combined in one large database with hundreds of cases. This database served as a pilaster for the design of a principal component analysis (PCA)-guided algorithm for prospective classification of individual tumor cells per case, which was then validated in a series of over 780 AL cases. Our results support the use of the ALOT database-guided analytical tools to harmonize diagnostic strategies and to facilitate fast, objective and reproducible diagnostics. In addition, these tools may be used to improve selection of markers in a robust manner for refined future WHO classification of AL.
Materials and methods
Acute leukemia patients
Patient characteristics are summarized in Table 1 and Supplementary Table 1. The institutional review board of each participating center approved this study and informed consent for study participation was obtained from each patient and/or his/her legal guardian.
Immunophenotyping
All samples were stained with the EuroFlow ALOT.8 Samples were stained and acquired according to the EuroFlow Standard Operating Procedure (SOP) and EuroFlow instrument settings.7 Appropriate instrument performance and laboratory procedures were confirmed by results obtained in the EuroFlow quality assessment rounds12 and by additional analyses (see Supplementary Information).
Study design
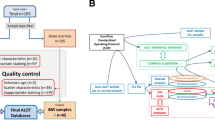
Data were collected in all participating EuroFlow centers. Resulting flow cytometry standard (.fcs) files were anonymized and analyzed using a standard profile in Infinicyt software (Cytognos, Salamanca, Spain), according to uniform gating instructions. Data (.fcs files and their corresponding analyzed.cyt files) were uploaded on a secured server (box-A), checked by a second expert (for correct acquisition, gating and fluorescence compensation), and if approved, subsequently transferred to box-C, together with all required annotations (Supplementary Figure 1). Atypical cases (for example, MPAL, AUL, blastic plasmacytoid dendritic cell neoplasms) were not transferred to box-C but to box-X. Data in box-C from all centers except two (Salamanca and Rotterdam) were used for building the ALOT database. The resulting ALOT database was validated using the remaining cases in box-C (Salamanca and Rotterdam cases) and atypical cases from box-X, as well as prospectively acquired new AL samples obtained in all participating centers for a total of 783 AL patients (47 T-ALL, 158 BCP-ALL, 522 AML, 55 MPAL, 1 AUL). Based on the (blinded) results, several cases were requested to be reviewed locally, taking into account not only the ALOT results, but also the information available from subsequent classification panels and genetics. Additional details are provided in the Supplementary Methods.
ALOT database
For construction of the ALOT database, available.cyt files from 747 patients were merged and grouped according to WHO-diagnosis in T-ALL, BCP-ALL and AML groups. After performing quality checks (see Results section), maximally 10 000 blast cells per case were exported from 656 cases and included in the ALOT database. The final ALOT database contained cases belonging to three disease categories (groups): T, B and Myeloid AL. Each group of cases was subsequently plotted in a balanced PCA plot,13 for comparisons of (i) each case versus each of the three database groups, (ii) each case versus all three groups at once, and/or (iii) individual cases versus combinations of two database groups, the latter two comparisons using pre-fixed (balanced) PCA plots.
ALOT database-guided analysis tool
An automated database-guided analytical algorithm and software tool was developed to classify individual leukemic cells from new patients in different categories, based on sequential and/or simultaneous comparison with cells from the AL patients in the database. Details about the algorithm can be found in Supplementary Data and Supplementary Figures 4-10. Briefly cells were classified as follows (Figure 1):
-
1
Typical cells, related to single lineage cells (T, B, or Myeloid), were defined as events falling within the 2 s.d. of a single AL group and outside the 2 s.d. of the two other groups, when comparing the testing case to each individual disease category independently in a PCA-guided analysis. They were represented in the Compass software tool as a single-color pointer directed to the appropriate disease category associated with the percentage of the corresponding cells.
-
2
Mixed cells were defined as cells that were not typical and fell outside the 2 s.d. of the dominant group when challenging the case with balanced supervised pairwise PCA-comparison of the two dominant groups. These cells were represented in the Compass tool as a dual-color arrow directed towards the mixed component associated with the percentage of cells.
-
3
Transitional cells were defined as virtually typical cells that fell within the 2 s.d. of the dominant group when challenging the case with balanced unsupervised pairwise PCA-comparison of the two dominant groups. These cells were represented in the Compass tool as a joint section (sector) in between the typical (that is, T, B, Myeloid) and mixed (T/B, T/M, B/M) components. By definition, transitional cells could not exist if no mixed component was identified.
-
4
Other cells were also identified, namely, triphenotypic (T/B/M) cells represented by a percentage in the middle of the Compass tool; and immature cells, represented as a gray circle whose diameter was proportional to the percentage of immature events. The classification system of these specific events is described in Supplementary Figures 2. Finally, unclassified cells included events that did not fulfill any of the criteria for any of the above described types of phenotype, and they were not represented in the Compass tool.
Schematic overview of the work process of the database-guided classification of leukemic cells from individual patients during the validation phase (Compass tool). The cells are classified as one of the indicated cell types based on the definitions in the second column, in the order as shown from top to bottom. Examples are shown in the right column, in which the three reference groups are represented in blue (T-ALL), green (BCP-ALL) and red (AML) (circles represent the mean of individual patients, the lines represent the 2 s.d. curves of the group), and the cells from the case to be classified are represented by the individual gray and black events. The black events indicate the events that fulfill the definition in the second column. Once a cell has been classified, it is removed from the subsequent steps. The different cell types are visualized in a final Compass, in which each cell type is represented in a specific manner (third column, see examples in Supplementary Information).
This database-guided analysis was implemented as the Compass tool in the Infinicyt 2.0 software and it is now available worldwide.
Statistical analyses
The probability of selecting the wrong antibody panel and make the wrong diagnosis by using only ALOT (expressed as p-values) and their corresponding 95% confidence intervals (95% CI), as well as the required sample sizes for a p-value <0.05 or <0.08, were calculated based on the number of cases evaluated and the actual mistakes made for all cases and per individual WHO diagnostic subgroup using a proportional statistical test (MATLAB software, Mathworks, Natick, MA, USA).
Results
Building of the EuroFlow ALOT database
Data from 747 ALOT cases were used for the database (165 T-ALL, 422 BCP-ALL, 160 AML cases). These cases first underwent a quality check, by evaluating MFI values of multiple markers on normal or malignant leukocytes. Cases with obviously atypical expression (n=81/747; 10.8%) due to technical errors/sample quality issues, were excluded from further analysis (Supplementary Figure 11 and Supplementary Table 2). The resulting 666 cases were merged into a single.fcs file. Using APS plots, with medians for each patient and SD curves for each patient group shown, cases outside the 2 s.d. curves were checked and excluded; this concerned 10 cases (1.5%). Therefore, the resulting EuroFlow ALOT database consisted of 656 cases (145 T-ALL, 377 BCP-ALL, 134 AML) (Figure 2).
Construction of the database and selection of cases. Schematic overview of the construction of the database: initially 747 cases were evaluated. After a first quality check 666 cases passed on to the next stage. These 666 cases were merged in a single file; 10 cases which felt out of the 2 s.d. curves were excluded. The remaining 656 cases entered the final ALOT database.
Contribution of markers in separating B, T and Myeloid
To evaluate the markers most contributing to the separation of T-ALL, BCP-ALL and AML cells, the principal components of the APS plot, with all three groups of patients, were analyzed (Supplementary Figure 12). The first principal component (x-axis) showed major contribution of CD7 (24%), CD19 (24%) and CyCD3 (18%) and particularly separates T-ALL from BCP-ALL. The second principal component (y axis) showed major contribution of CyMPO (21%), FSC (17%), SSC (14%) and particularly separated AML from BCP-ALL and T-ALL.
Database-guided classification of AL patients
The resulting ALOT database and the newly developed database-guided analysis tool were validated on 783 cases (47 T-ALL, 158 BCP-ALL, 522 AML; 1 AUL, 55 MPAL). All cases were analyzed using the database-guided analysis tool and the results were compared with the final WHO2008 diagnosis, as established by the individual centers. Based on the database-guided analysis, patients were categorized as follows:
-
Typical T, B or Myeloid (without mixed cells): the database-guided analysis shows typical cells for only one lineage (either T, B or Myeloid) and there are <1% mixed or transitional cells.
-
Typical T, B or Myeloid for only one lineage plus mixed cells: the database-guided analysis shows>10% typical cells (either T, B or Myeloid), and >1% of mixed cells (T/M, T/B, M/B)
-
Atypical: the database-guided analysis shows <10% of typical cells (either T, B or Myeloid), irrespective of the percentage of mixed cells.
-
Mixed lineage: the database-guided analysis shows typical cells in at least two lineages (either T+B, T+M, M+B, or T+B+M), either with or without mixed or transitional cells.
The individual data for the cases in the various categories are shown in Supplementary Table 4. Overall, the distribution of the 783 cases over these categories was 535 (68%), 170 (22%), 9 (1%) and 69 (9%), respectively, and their distribution over the different WHO categories is shown in Table 2. In 40/47 (85%) T-ALL, 153/158 (97%) BCP-ALL and 498/522 (95%) AML patients, the database-guided analysis classified cells as typical T, B or Myeloid, respectively. Furthermore, in 38/55 (69%) of MPAL-classified patients the database-guided analysis showed typical cells in at least two lineages (‘mixed lineage cases’) and in an additional 10/55 (18%) cases the database-guided analysis indicated either large numbers of mixed cells (>10%) or very low numbers of typical cells (<10%), resulting in 87% of MPAL cases well recognized.
Detailed evaluation of typical B, T or Myeloid cases (without mixed cells)
Overall, 395/535 typical cases were classified as typical AML. The average percentage of cells (±s.d.) defined as typical Myeloid was 95.4±9.1%. In only four cases the percentage of typical cells was <50%, but always >10%. Review of these cases showed one acute promyelocytic leukemia, one secondary AML, one t(8;21)-positive AML and one AML case which weakly expressed CD19 and partially expressed CyMPO. Two cases classified as typical AML by the database-guided analysis were originally diagnosed with an MPAL (see Supplementary Table 8). Blinded local review resulted in a diagnosis of AML with aberrant B-cell markers for both cases.
In turn, 110/535 cases were classified as typical BCP-ALL. The average percentage of cells (±s.d.) defined as typical B was 96.6%±6.7%; in all cases the percentage was >50%. Two cases were initially diagnosed as MPAL. Blinded local review resulted in a diagnosis of BCP-ALL with aberrant weak CyMPO expression for both cases (see Supplementary Table 8).
Finally, 30 cases were classified as typical T-ALL. The average percentage of cells (±s.d.) defined as typical T was 95.0%±9.9%; in all cases the percentage was >50%. All cases were diagnosed as T-ALL.
Detailed evaluation of Typical T, B or Myeloid with mixed cells cases
In total, 170 cases (10 T-ALL 45 BCP-ALL, 105 AML and 10 MPAL) were categorized as T, B or Myeloid with mixed populations, and had ⩾10% of events defined as typical T, B or Myeloid (average±s.d.: 71±23% for T-ALL 67±24% for BCP-ALL and 59±27% for AML). By definition, all cases also had >1% of mixed cells; on average 9±10% were within these mixed populations (T/B: 16±16%; T/M: 7±7%; M/B: 10±10%; TMB: 10±13%). These cases were further subdivided based on the percentage of cells in the mixed population. Fifty-five cases had ⩾10% mixed cells and of these 7 (13%) were diagnosed as MPAL. In contrast, only 3/115 (3%) cases with <10% mixed cells were diagnosed with an MPAL. After blinded review of these three MPAL cases, one was categorized as BCP-ALL with aberrant weak CyMPO+,14 whereas the diagnosis of MPAL was maintained in the other two cases, due to the presence of a major myeloid population and a small B-cell population (see Supplementary Table 8). In the latter cases, the two populations may better be analyzed separately in the database-guided analysis.
Detailed evaluation of atypical cases
In seven cases the database-guided analysis defined <10% of cells as typical (either T, B or Myeloid). Three cases were diagnosed as MPAL (see Supplementary Table 8). Case 4 showed typical Myeloid cells and mixed T/M cells and was diagnosed with a GATA1+ Down syndrome (DS)-AML. Case 5 was diagnosed with a second relapse BCP-ALL but the database-guided analysis suggested many mixed M/B cells; central review showed large cells with a clearly defined CD19+/CyCD79a+/CD34+/weak CyMPO+ MPAL immunophenotype. Case 6 was diagnosed as T-ALL; central and local review suggested an MPAL T/M (CyCD3+/weak CyMPO+). Case 7 was diagnosed as T-cell malignancy with a CD3+/CyCD3+/TCRgd+/CD34-/CD117-/CD1a-/CD10-/TdT-/CD4-/CD8-/CD2+/CD5-/heterogeneous/CD7+/CD45+ immunophenotype: due to the absence of immature markers likely a mature T-cell malignancy.
In two patients, the database-guided analysis did not show typical cells at all. In one case the database-guided analysis assigned most events as T/M cells (6%) and transitional T>T/M cells (86%). Central review showed that the leukemic cells were CD34+/weak CD45+/CyMPO-/CD19-/CyCD79a-/partially CD7+/partially CyCD3+. Based on additional immunostainings, this case was diagnosed as early-T-cell precursor (ETP) ALL. The second case was CD34-/weak CD45+/CyCD3+/SmCD3-/strong CD7+/CD19-/CyCD79a-/weak CyMPO+; all events were defined unclassified by the database-guided analysis. Further evaluations showed TdT-positivity and a double NOTCH1 mutation; the case was diagnosed as T-ALL.
Clearly, most of these cases were indeed atypical and required special attention and subsequent staining of multiple classification panels.
Detailed evaluation of mixed lineage cases
In 69 cases, the database-guided analysis classified cells as typical in two or more lineages, suggesting truly mixed-lineage phenotype AL. Indeed, in 38 cases (55%) diagnosis of MPAL was finally made (see Figure 3 for an example). One case (1%) was diagnosed as an AUL, the database-guided analysis indicated to typical Myeloid and typical B cells in addition to mixed T/M, M/B, and TBM cells and about one-third of transitional M>M/B cells. Of the remaining 30 cases (43%; see Supplementary Table 8), most had typical Myeloid and T cells; review of these cases frequently showed a CyMPO-/CD7+ immunophenotype (AML with minimal differentiation), generally also TdT was positive in these cases (Figure 4). Other cases showed an atypical immunophenotype, including one ETP-ALL, two blastic plasmacytoid dendritic cell neoplasms, one t(9;22) and two MLL-positive cases. Finally, in a few cases one of the populations defined as typical seemed to be due to non-purity of the gated blast cell population, resulting in contamination with for example dendritic cells or normal B-cell precursors. Such contaminations of the leukemic cell gate, generally could well be identified by back-gating using the database-guided analysis plots.
Example of a patient with an MPAL. The leukemic cells (in red) are dim CD45+/CyCD3-/SmCD3-/partially CyMPO+/partially CD34+/partially CD19+/partially CyCD79a+. The database-guided analysis showed a main pointer to Myeloid (34%) and B (52%), indeed suggesting an MPAL. The overall immunophenotype was partially CD34+/partially CD117+/weak HLADR+/weak CD45+/CD13+/CD33+/CD11b-/CD14-/CD15-/CD16-/CD36-/CD64-/CD56-/partially TdT+/heterogeneous CD123+/CD66c+/CyIgM-/partially CD10+/IgM-/partially CD22+/CD20-. Morphology showed 90% blasts, Sudan black positive in 5% of cells. Cytogenetics showed 46,XX,t(9;22)(q34;q11), BCR-ABL1-positive. According to WHO2008 the patient was diagnosed as MPAL (myeloid/B) with t(9;22). Normal T-cells (dark blue), NK cells (middle blue), mature B-cells (green), eosinophils (light blue), granulocytes (purple), monocytes (orange) and immature erythroid cells (brown) are shown as well.
Example of a patient with an AML with minimal differentiation. The overall immunophenotype was CD34+/CD117+/weak HLADR+/weak CD45+/CD13-/partially CD33+/CD11b-/CD14-/ CD15-/CD16-/partially TdT+/CD36-/CD64-/CD56-/CD4-/heterogeneous CD123+/CD2-/CD5-. Morphology showed an AML, FAB-M0. Cytogenetics were 46,XY, t(1;11)(q21;q23). According to WHO2008 the patient was diagnosed as AML (with minimal differentiation). Normal T-cells (dark blue), NK cells (middle blue), mature B-cells (green), eosinophils (light blue), granulocytes (purple), monocytes (orange) and immature erythroid cells (brown) are shown as well.
Algorithm for selection of appropriate classification panels
We propose the algorithm shown in Figure 5 for selection of the appropriate classification panels. This algorithm directly guides the user to the appropriate single classification panel in 84 of cases. In 16% of cases the database-guided analysis indicates that there may be a mixed population and that the expert should carefully evaluate the data and select the appropriate classification panel(s). Using this algorithm, in over 99% of cases the right classification panel(s) will be selected, in only two cases (0.3% of all patients) the database-guided analysis directed towards an AML panel whereas diagnosis of MPAL was made based on the presence of a subset of cells that also expressed B-cell markers. Based on the validation series results, the p-values for the total cases and for each WHO diagnostic subgroup separately were: total cases: 0.004 (95% CI: <0.001–0.009); AML: 0.004 (<0.001–0.004); BCP-ALL: <0.001 (<0.001–<0.001); T-ALL: <0.001 (<0.001–<0.001); MPAL: <0.001 (<0.001–<0.001). Based on the wrong decisions detected in the validation series the estimated number of cases required for an error of <5% (p-value <0.05) per WHO diagnostic subgroup would be lower than 10.
Proposed algorithm for application of the Compass tool results. Cases in which the Compass shows a main pointer towards B, T, or Myeloid (with >10% of events) and that have no mixed events or <10% mixed events can be stained immediately with the classification panel of the main pointer. This accounts for 84% of cases (n=650); of these only two (0,3%) were classified as MPAL after review. All other cases (16% of total; n=133) need manual selection of the right classification panel; 36% of these were diagnosed as MPAL.
Although the ALOT tube and database are not meant for making a final diagnosis but for guiding towards the appropriate classification panel, the database-guided analysis provided data comparable to the final WHO diagnosis already in >93% of cases (p=0.046 (95%CI: 0.022–0.070; see Supplementary Information for statistical details).
Discussion
Flow cytometry is a frontline tool for orientation towards the right diagnosis and the appropriate treatment protocol in AL. However, no robust individual marker is sufficient to correctly identify each major disease category and consequently, interpretation requires the integration of complex patterns of immunophenotypes. In addition, the inherent biological variability of the disease makes the overall picture even more complex, with overlapping (for example, AML1-ETO AML co-expressing B-cell markers), intermingled (for example, MPALs) or even unclassifiable (AUL) immunophenotypes. Hence, it is recognized that immunophenotyping remains highly sensitive to technical procedures, including expert-based identification of the leukemic cells, and strongly subjective and dependent on the expertise of laboratory professionals for optimal interpretation of results. Within the EuroFlow consortium, we previously reported the ALOT tube to efficiently direct towards the appropriate classification panel for full characterization of AL. It included the major markers for frontline screening of myeloid, B and T-cell lineages, and also for detection of potentially more complex immunophenotypes, such as MPALs and AULs. In this study, we built an ALOT database and a PCA-based algorithm and software tool for further supporting more objective and reproducible local selection of the appropriate classification panel(s).
Construction of the reference database required application of standardized instrument settings, staining protocols and antibody panels in all participating laboratories.7, 8 Cases with clearly atypical expression of certain markers were excluded from the database; such atypical expression was most likely related to technical problems or sample quality issues, although some biological variability could not be ruled out in a few cases. The relatively large number of excluded cases (12%) indicates that, despite standardization, results are still not always comparable and that stringent quality assurance programs per sample are required.12 We aimed to build the reference database with typical BCP-ALL, T-ALL and AML cases (656 cases in total). Thus, MPAL cases were initially not included but temporarily stored aside and finally evaluated in the validation cohort.
The ALOT tube and database are not meant for making a final diagnosis but for guiding towards the appropriate classification panel. Using the here-proposed algorithm, the right classification panel(s) will be selected in >99% of cases. It should be noted that the classification panels also contain ‘cross-lineage’ markers (CD19, CD22 and CD10 in AML panel, CD13, CD33 and CD117 in BCP-ALL and T-ALL panels)7 that will facilitate right interpretation of the final data, even if only a single classification panel is originally selected. Nevertheless, given the composition of the ALOT tube (including most lineage-defining markers) the database-guided tool already provides data comparable to the final WHO classification in >93% of cases.
Prospective comparison of blast cells from individual AL cases against the ALOT reference database using the ‘big data’ analytical strategies here proposed, guides/helps the expert on the interpretation of the overall immunophenotypic pattern of leukemic cells in the diagnosis workup of hematological malignancies. The advantage of the procedure is the capability of the algorithm to objectively analyze all single leukemic events and to classify each of them individually. As such, it provides a snapshot of the exact composition of the leukemic bulk by describing all cellular subsets that reflect the intrinsic phenotypic heterogeneity of AL. This single-cell analysis relies on accurate simultaneous measurements of MFIs from eight different markers plus two light scatter parameters. This is obviously more objective, and in sharp contrast with the expert-based interpretation, which takes into account an arbitrary categorical classification of negative versus positive and dim versus bright patterns of marker expression for the dominant leukemic population,15 with no clear insight into the phenotypic complexity of the total tumor load. As a consequence, the algorithm is more likely to discern minor immature cells or leaky phenotypes that are ignored by the expert-based analysis. In that sense, the computer tools draw the attention of the expert to possibly relevant subcomponents of the leukemia, at the same time they fully allow for the parallel conventional expert-based diagnostic interpretation. This may be of particular relevance to prevent from missing MPALs because of minor components, as found here for a few discrepant cases. In line with this, one could hypothesize that these minor components may be underestimated, possibly providing also a rationale for switching-lineage leukemias.16 From a nosological perspective, this further raises the question about the potential relevance of the ability to dissect with precision the phenotypic composition of the leukemic population. Thus, we provide here an objective tool to decipher the phenotypic complexity of the leukemic population that may meet clinical correlations, and raises the possibility to revisit the criteria of the WHO classification in the near future.
In this regard, the PCA-based algorithm applied to the three typical AL groups of the reference database also highlighted the relative contribution of each marker. Regarding the separation between the three disease categories, CD7 had a major contribution besides CD19 and CyCD3, and in fact CD7 was the most contributing parameter. This may sound counter-intuitive at first glance given the fact that CD7 expression can also be seen in AML. As a consequence, this approach could be used to further improve the WHO classification and one should consider including the intensity of CD7 expression of individual leukemic cells within a patient, in future classification schemes for AL. As for the atypical cases, notably MPALs and AULs, these are considered as more difficult diagnoses. Despite the criteria defined by the WHO classification, the interpretation remains subjective, giving rise to frequent file sharing between experts to consensually state the diagnosis. This automated approach, once applied together with standardized protocols and reagent panels, provides an innovative and reproducible approach for robust delineation of these disease categories. However, our strategy is extremely sensitive to the quality of the selection criteria (that is, gates) used to identify the leukemic cells, which might also benefit from the automated gating strategies recently developed by the EuroFlow consortium.6 Thus, in the near future, databases with ALOT data from normal subjects and patients may allow automated gating of bone marrow and peripheral blood normal and leukemic cell populations,6 thereby making the whole analyses even more objective by avoiding contamination of the leukemic cells gates with normal residual cellular events that may responsible for false alarms of mixed lineage components.
In conclusion, here we show that building of large multicenter databases of flow cytometry immunophenotypic data is feasible, if instrument settings, staining protocols and antibody panels are well-standardized. The combination of such database with database-guided analytical tools may support interpretation of flowcytometric data and guide the user towards appropriate subsequent antibody panels. The ALOT database (656 AL patients) and database-guided analytical tools were validated in >780 patients and our results show that both typical and atypical (mainly MPAL) cases could be recognized correctly in >99% of cases. Thus, usage of the database-guided analysis may contribute to more objective interpretation of immunophenotypic data and improved diagnostics. This paves the way for improved database-guided and computer-supported reproducible classification of single leukemic cells from individual patients, supervised by an expert, also in other hematological malignancies and non-hematological diseases.
References
Borowitz MJ, JKC Chan B lymphoblastic leukaemia/lymphoma. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW (eds). WHO classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. International Agency for Research on Cancer: Lyon, 2008, pp 168–175.
Borowitz MJ, JKC Chan T lymphoblastic leukaemia/lymphoma. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. International Agency for Research on Cancer: Lyon, 2008, pp 176–178.
Borowitz MJ, Bene MC, Harris NL, Porwitt A, Matutes E. Acute leukaemias of ambiguous lineage. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. International Agency for Research on Cancer: Lyon, 2008, pp 150–155.
Mejstrikova E, Volejnikova J, Fronkova E, Zdrahalova K, Kalina T, Sterba J et al. Prognosis of children with mixed phenotype acute leukemia treated on the basis of consistent immunophenotypic criteria. Haematologica 2010; 95: 928–935.
van den Ancker W, Terwijn M, Westers TM, Merle PA, van Beckhoven E, Drager AM et al. Acute leukemias of ambiguous lineage: diagnostic consequences of the WHO2008 classification. Leukemia 2010; 24: 1392–1396.
Flores-Montero J, Sanoja-Flores L, Paiva B, Puig N, Garcia-Sanchez O, Bottcher S et al. Next Generation Flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017; 31: 2094–2103.
Kalina T, Flores-Montero J, van der Velden VH, Martin-Ayuso M, Bottcher S, Ritgen M et al. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia 2012; 26: 1986–2010.
van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012; 26: 1908–1975.
Flores-Montero J, Flores LS, Paiva B, Puig N, Garcia-Sanchez O, Bottcher S et al. Next generation flow (NGF) for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017; 31: 2094–2103.
Theunissen P, Mejstrikova E, Sedek L, van der Sluijs-Gelling AJ, Gaipa G, Bartels M et al. Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood 2017; 129: 347–357.
Han K, Kim Y, Lee J, Lim J, Lee KY, Kang CS et al. Human basophils express CD22 without expression of CD19. Cytometry 1999; 37: 178–183.
Kalina T, Flores-Montero J, Lecrevisse Q, Pedreira CE, van der Velden VH, Novakova M et al. Quality assessment program for EuroFlow protocols: summary results of four-year (2010-2013) quality assurance rounds. Cytometry A 2015; 87: 145–156.
Costa ES, Pedreira CE, Barrena S, Lecrevisse Q, Flores J, Quijano S et al. Automated pattern-guided principal component analysis vs expert-based immunophenotypic classification of B-cell chronic lymphoproliferative disorders: a step forward in the standardization of clinical immunophenotyping. Leukemia 2010; 24: 1927–1933.
Steiner M, Attarbaschi A, Dworzak M, Strobl H, Pickl W, Kornmuller R et al. Cytochemically myeloperoxidase positive childhood acute leukemia with lymphoblastic morphology treated as lymphoblastic leukemia. J Pediatr Hematol Oncol 2010; 32: e4–e7.
Pedreira CE, Costa ES, Lecrevisse Q, van Dongen JJ, Orfao A, EuroFlow C . Overview of clinical flow cytometry data analysis: recent advances and future challenges. Trends Biotechnol 2013; 31: 415–425.
Slamova L, Starkova J, Fronkova E, Zaliova M, Reznickova L, van Delft FW et al. CD2-positive B-cell precursor acute lymphoblastic leukemia with an early switch to the monocytic lineage. Leukemia 2014; 28: 609–620.
Acknowledgements
The research was performed within the EuroFlow Consortium, which started with an EU-FP6 grant (LSHB-CT-2006-018708) and obtained sustainability by protecting and licensing intellectual property, thereby obtaining royalties, which are exclusively being used for supporting the EuroFlow research program (chairmen: JJMvD and AO). LS, JB and TS were supported by STRATEGMED3/304586/5/NCBR/2017 PersonALL grant of the Polish National Center for Research and Development. ESC acknowledges FAPERJ, Rio de Janeiro, Brazil (E26/110.105/2014; E26/102.191/2013) and Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPQ of Brazil (400194/2014-7). AO, SM and AL acknowledge the Instituto de Salud Carlos III (MINECO, Madrid, Spain) for the DTS15/00119, and CIBERONC-FEDER-CB16/12/00400 grants. EM, OH, TK and MN were supported by Ministry of Health grant number 15-28525A and NPU LO1604. The research for this manuscript was in part performed within the framework of the Erasmus Postgraduate School Molecular Medicine.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
JJMvD: contract research for Roche, Amgen and BD Biosciences. VHJvdV: contract research for Roche, Amgen, Pfizer, Janssen and BD Biosciences; consultancy fees: Celgene. TS: a member of a Scientific Advisory Board for JazzPharma, Novartis; reimbursement for attending symposia organized by CSL Behring, Novartis, Pfizer, Alexion, Shire and JazzPharma. GEG and MMA are employees of Cytognos SL, Salamanca, Spain. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Lhermitte, L., Mejstrikova, E., van der Sluijs-Gelling, A. et al. Automated database-guided expert-supervised orientation for immunophenotypic diagnosis and classification of acute leukemia. Leukemia 32, 874–881 (2018). https://doi.org/10.1038/leu.2017.313
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.313
This article is cited by
-
LncRNAs serve as novel biomarkers for diagnosis and prognosis of childhood ALL
Biomarker Research (2021)
-
Ferritin-based targeted delivery of arsenic to diverse leukaemia types confers strong anti-leukaemia therapeutic effects
Nature Nanotechnology (2021)
-
Monitoring minimal/measurable residual disease in B-cell acute lymphoblastic leukemia by flow cytometry during targeted therapy
International Journal of Hematology (2021)
-
Automated identification of leukocyte subsets improves standardization of database-guided expert-supervised diagnostic orientation in acute leukemia: a EuroFlow study
Modern Pathology (2021)
-
A novel flow cytometric method for enhancing acute promyelocytic leukemia screening by multidimensional dot-plots
Annals of Hematology (2019)