Abstract
Mutations in the interleukin-7 receptor (IL7R) or the Janus kinase 3 (JAK3) kinase occur frequently in T-cell acute lymphoblastic leukemia (T-ALL) and both are able to drive cellular transformation and the development of T-ALL in mouse models. However, the signal transduction pathways downstream of JAK3 mutations remain poorly characterized. Here we describe the phosphoproteome downstream of the JAK3(L857Q)/(M511I) activating mutations in transformed Ba/F3 lymphocyte cells. Signaling pathways regulated by JAK3 mutants were assessed following acute inhibition of JAK1/JAK3 using the JAK kinase inhibitors ruxolitinib or tofacitinib. Comprehensive network interrogation using the phosphoproteomic signatures identified significant changes in pathways regulating cell cycle, translation initiation, mitogen-activated protein kinase and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT signaling, RNA metabolism, as well as epigenetic and apoptotic processes. Key regulatory proteins within pathways that showed altered phosphorylation following JAK inhibition were targeted using selumetinib and trametinib (MEK), buparlisib (PI3K) and ABT-199 (BCL2), and found to be synergistic in combination with JAK kinase inhibitors in primary T-ALL samples harboring JAK3 mutations. These data provide the first detailed molecular characterization of the downstream signaling pathways regulated by JAK3 mutations and provide further understanding into the oncogenic processes regulated by constitutive kinase activation aiding in the development of improved combinatorial treatment regimens.
Similar content being viewed by others
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive leukemia that is common in children and adolescents. Long-term childhood ALL survival rates have improved significantly following refined chemotherapeutic treatment regimens; however, these are associated with substantial acute and long-term side effects.1 Recently, Janus kinase 3 (JAK3) mutations were identified in 16% of T-ALL cases2 and it is now known to be sufficient to drive T-ALL development in mice using an in vivo bone marrow transplant model.3 JAK3 is a non-receptor tyrosine kinase and functions in class I cytokine receptor complexes through binding of the common γ chain (IL2RG). JAK3 binds the IL2RG that forms heterodimers with other receptors such as the IL7Rα chain, which in turn binds JAK1. Under normal conditions, ligand binding to the receptor complex activates cytokine signaling through cooperative JAK1/JAK3 phosphorylation. In this active conformation, they phosphorylate downstream targets including STAT5. In addition to canonical STAT5 activation, increasing evidence shows that the interleukins (ILs) can activate additional signaling pathways in T-cells. For example, a phosphoproteomic approach following 5 min stimulation with IL2 and IL15 found recruitment and phosphorylation of the SHC–GRB2–SOS complex at the cytokine receptor which then activates the canonical mitogen-activated protein kinase (MAPK) pathway.4 Likewise, very early work found that IL7 stimulation leads to JAK3-mediated association and phosphorylation of the p85 subunit of the PI3-kinase.5
The recent identification of activating JAK3 mutations in T-ALL cases shows promising therapeutic potential.6, 7, 8 Indeed, small molecules targeting JAK family members are in development or are already in use for the treatment of several diseases which could be repurposed for T-ALL.9 However, detailed functional analysis of the different JAK3 mutations has found these have different dependencies on members of the IL7R complex for their ability to cause cellular transformation.3, 10 For example the JAK3(M511I) mutation requires the presence of both JAK1 and the IL2RG for cellular transformation, whereas the JAK3(L857Q) can also signal independent from the JAK1/IL2RG complex.3, 10 Therefore, the pathways activated downstream of different JAK3 mutations in additional to canonical STAT5 activation, may not only differ from wild-type cytokine activation of JAK3, but also between the different JAK3 mutations themselves. Hence, the detailed molecular characterization of the oncogenic JAK3 pathways based on the exact mutation present will aid in the development of improved treatment strategies.
Accordingly, we have used quantitative assessment of the phosphorylation status of proteins downstream of mutant JAK3(L857Q) and JAK3(M511I) using transformed Ba/F3 cells treated with the JAK1/JAK3-selective inhibitors ruxolitinib or tofacitinib, or a vehicle control. Our data have mapped associations between mutant JAK3 signaling and multiple components of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT and MAPK pathways, as well as to critical components of the cell cycle machinery, anti-apoptotic constituents, RNA metabolism and epigenetic regulators.
Materials and methods
Expression plasmids
JAK3 mutant sequences were synthesized by GenScript (Piscataway, NJ, USA). All constructs were cloned into the murine stem cell virus–green fluorescent protein vectors.
Cell culture, virus production and retroviral transduction
Cell culture, virus production and retroviral transduction were performed as previously described.3
Quantitative phosphoproteomics
Discovery
Three independent sets of Ba/F3 cells expressing mutant human JAK3(L857Q) were cultured as described previously3 and treated with 500 nM tofacitinib, ruxolitinib or dimethylsulfoxide for 90 min (3 × biological). Cell pellets were prepared as previously described.11 Membranes were enriched by ultra-centrifugation12 and proteins dissolved in v/v 6 M urea/2 M thiourea. Proteins were then reduced, alkylated and digested as previously described.11 Lipids were precipitated from membrane peptides using formic acid and quantitated (Qubit protein assay kit, Thermo Fisher Scientific, Carlsbad, CA, USA). One hundred micrograms of membrane and soluble peptides from each of the nine samples were individually labeled using tandem mass tags (TMT-10plex 2 × kits, Thermo Fisher Scientific, Bremen DE, Germany) and mixed with a 1:1 ratio.13 Phosphopeptides were isolated from the proteome using titanium dioxide and immobilized metal affinity chromatographybefore offline hydrophilic interaction liquid chromatography (LC).14 LC tandem mass spectrometry (MS/MS) was performed on 24 mono-phosphopeptide enriched hydrophilic interaction LC fractions, 12 membrane and 12 soluble enriched fractions, as well as multi-phosphorylated peptides enriched from both using a Q-Exactive Plus hybrid quadrupole-Orbitrap MS system (Thermo Fisher Scientific, Bremen, Germany) coupled to a Dionex Ultimate 3000RSLC nanoflow HPLC system (Thermo Fisher Scientific). Samples were loaded onto an Acclaim PepMap100 C18 75 μm × 20 mm trap column (Thermo Fisher Scientific) for pre-concentration and online desalting. Separation was then achieved using an EASY-Spray PepMap C18 75 μm × 500 mm column (Thermo Fisher Scientific), employing a linear gradient from 2 to 32% acetonitrile at 300 nl/min over 120 min. Q-Exactive Plus MS System (Thermo Fisher Scientific) was operated in full MS/data-dependent acquisition MS/MS mode (data-dependent acquisition). The Orbitrap mass analyzer was used at a resolution of 70 000, to acquire full MS with an m/z range of 390–1400, incorporating a target automatic gain control value of 1e6 and maximum fill times of 50 ms. The 20 most intense multiply charged precursors were selected for higher-energy collision dissociation fragmentation with a normalized collisional energy of 32. MS/MS fragments were measured at an Orbitrap resolution of 35 000 using an automatic gain control target of 2e5 and maximum fill times of 110 ms.
Parallel reaction monitoring
Phosphopeptides were loaded onto an Acclaim PepMap100 C18 75 μm × 20 mm trap column (Thermo Fisher Scientific) for pre-concentration and online desalting, then separated using EASY-Spray PepMap C18 75 μm × 250 mm column (Thermo Fisher Scientific). An optimized stepped 82 min gradient was employed (5 to 22 to 35%). Each sample was run in data-dependent acquisition mode (described above) to evaluate phosphopeptide enrichment efficiency (93–97%) and loading. Parallel reaction monitoring (PRM) was performed using optimized methods for collision energy, charge state and retention times at a resolution of 17 500, automatic gain control of 2e5, maximum fill times of 90 ms and 1.6 m/z isolation window.
Data analysis
Database searching of all.raw files was performed using Proteome Discoverer 2.1 (Thermo Fisher Scientific). Mascot 2.2.3 and SEQUEST HT were used to search against the Swiss_Prot, Uniprot_mouse database (24910 sequences, downloaded 19 April 2016). Database searching parameters included up to two missed cleavages, to allow for full tryptic digestion, a precursor mass tolerance set to 10 p.p.m. and fragment mass tolerance 0.02 Da. Cysteine carbamidomethylation was set as a fixed modification while dynamic modifications included oxidation (M), phospho (S/T), phospho (Y) and TMT6plex (modification designated for TMT10plex). Interrogation of the corresponding reversed database was also performed to evaluate the false discovery rate of peptide identification using Percolator on the basis of q-values, which were estimated from the target-decoy search approach. To filter out target peptide spectrum matches over the decoy-peptide spectrum matches, a fixed false discovery rate of 1% was set at the peptide level. The experimental workflow is shown in Figure 1. PRM data files were processed with PD 2.1 to generate spectral libraries and uploaded into Skyline (MacCoss Lab, University of Washington), which was then used to identify peptide chromatogram peaks. Peptide quantification was performed by quantifying area under the curve of the MS2 extracted ion chromatogram within Skyline.15 Results were normalized to the spiked-in heavy-labelled peptide to account for variations in sample injection. MS raw data is available in the Massive public repository (PXD007046).
Experimental workflow. Three independent sets of Ba/F3 cells expressing mutant human JAK3 (L857Q) were cultured and treated with 500 nM tofacitinib, ruxolitinib or dimethylsulfoxide (DMSO) for 90 min (biological and technical replicates). Cells were digested with Typ/Lys-C mix. Peptides from each of the nine samples were labeled using tandem mass tags (TMT-10plex, Thermo Fisher Scientific) and mixed 1:1. Phosphopeptides were isolated and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed using a Q-Exactive Plus hybrid quadrupole-Orbitrap MS system (Thermo Fisher Scientific). Database searching of all.raw files was performed using Proteome Discoverer 2.1 (Thermo Fisher Scientific). Mascot 2.2.3 and SEQUEST HT were used to search against the Swiss_Prot, Uniprot_mouse database.
In silico analysis of phosphosites abundance and pathway prediction
Quantitative phosphoproteomic differences across treatments were clustered by unique phosphosites (Cluster3, Stanford University, Palo Alto, CA, USA) and examined using heatmaps (Java Treeview, Stanford University) to visualize trends and consistency (as described16) in phosphopeptide abundance across treatment groups. Ingenuity Pathway Analysis software (version 8.8, Ingenuity Systems, Redwood City, CA, USA) together with the Core Analysis function was used to identify pathways regulated by the JAK3(L857Q) mutation.
Western blotting
Cells were lysed in cold lysis buffer containing 5 mM NA3VO4, protease inhibitors (Complete, Roche, Bazel, Switzerland) and PhosSTOP (Roche). The proteins were separated on NuPAGE NOVEX Bis-Tris 4 to 12% gels (Invitrogen, Carlsbad, CA, USA). Western blot analysis was performed using antibodies against JAK3 (Cell Signaling, Danvers, MA, USA 3775), phospho-STAT5 (Y694) (Cell Signaling 9359); STAT5 (Invitrogen 335900), phospho-RanBP3 (Cell Signaling 9380); phospho-4E-BP1 (Cell Signaling 2855); pERK1/2 (Cell Signaling 9101), ERK1 (Santa Cruz, Dallas, TX, USA, Sc-93), pMEK1/2 (Cell Signaling 9154), MEK1/2 (Cell Signaling 9126) and β-actin (Sigma-Aldrich A1978); secondary antibodies were conjugated with horseradish peroxidase (GE Healthcare, Chicago, IL, USA). Bands were visualized using a cooled charge-coupled device camera (ImageQuant LAS-4000; GE Healthcare).
Inhibitor treatment in Ba/F3 cells
Ba/F3 cells (www.dsmz.de) were cultured and transduced as described.3 Cells were confirmed mycoplasma negative. Ba/F3 cells were seeded in 96-well plates (1x105 cells/ml) and treated with compound or vehicle (dimethylsulfoxide). A quantitative evaluation of proliferation was done after 24 h using ATPlite (PerkinElmer, Waltham, MA, USA) and measured on the VICTOR X4 Reader (PerkinElmer).
Ex vivo treatment of primary T-ALL samples
The mutational background of T-ALL samples used in this work is shown in Supplementary Table S1. All human T-ALL samples were collected with informed patient consent and with approval of the UZ Leuven ethics committee. T-ALL cells were expanded in vivo by injection in 8 weeks old female NSG mice. After 2–3 months, human T-ALL cells were isolated from the spleen, seeded in 96-well plates (5 × 105 cells/well) and incubated with vehicle (dimethylsulfoxide) or inhibitor. Cell viability was assessed 48 h after adding the compounds by using the Guava easyCyte Flow Cytometer (Millipore, Overijse, Belgium). CompuSyn was used to calculate the combination index (CI). All animal experiments were approved by the KU Leuven ethics committee.
Annexin V/PI staining and flow cytometry analyses
For apoptosis staining the fluorescein isothiocyanate Annexin V apoptosis detection kit with propidium iodide was used as per the manufacturer’s instructions (Biolegend, Dedham, MA, USA). Briefly, 500 000 cells of each condition were stained for 20 min. Staining cells were washed in 1 × in phosphate-buffered saline and data were acquired on the MACSQuant VYB flow cytometer (Miltenyi Biotec, Cambridge, MA, USA). For phospho-flow analysis, cells were fixed with Inside Fix and permeabilisation buffer (Miltenyi Biotec), and stained with an APC-labeled antibody against phospho-STAT5 (eBioscience, San Diego, CA, USA). Cells were analyzed using the FACSVerse (BD, Bedford, MA, USA). All FACS data analysis was carried out using FlowJo sotware (Tree Star, Ashland, OR, USA).
In vivo drug treatment of primary T-ALL sample
Patient sample 389E was injected into the lateral tail vein of 28 female NSG mice. Leukemia burden was assessed by hCD45+ staining in the peripheral blood on day 19 post injection. Mice were ranked from highest to lowest hCD45 and then sequentially placed in four treatment groups. Animal weight distribution was also assessed to determine equivalence between all groups. Ruxolitinib and ABT-199 were administered daily by oral gavage for 14 consecutive days, vehicle was given as placebo. No preliminary data were available to perform a statistical power analysis. The investigators were not blinded to the treatment groups.
Results
JAK3 pathway network enrichment analysis
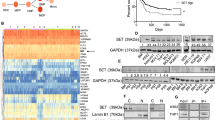
To determine the signal transduction pathways downstream mutant JAK3, Ba/F3 cells transformed by the JAK3(L857Q) mutation (signaling partially independent from JAK1) were initially screened using unbiased global quantitative phosphoproteomic profiling. This approach simultaneously determined the identity, phosphorylation status and relative abundance of proteins isolated from three biological replicates following JAK1 inhibition (ruxolitinib) or JAK3 inhibition (tofacitinib) (Figure 1). Deep phosphoproteomic coverage was achieved by employing a multidimensional strategy incorporating protein pre-fractionation and sequential phosphopeptide enrichment techniques to enrich for mono- and multi-phosphorylated peptides, coupled to hydrophilic interaction LC, to reduce sample complexity before high-resolution tandem MS. A total of 6036 unique proteins and 2070 unique phosphoproteins, associated with ~5400 unique phosphorylation sites were quantitatively sequenced across all 9 samples (Figures 2a–c and Supplementary Table S2). Accurate site-specific assignment of phosphorylation was achieved using phosphoRS17 using a false discovery rate of 1% followed by manual interrogation for all peptides that were significantly altered after JAK inhibition. The ratio of phosphosites identified were 82.5% serine: 14.9% threonine: 2.6% tyrosine.
Quantitative phosphoproteomic analysis of mutant JAK3 L857Q Ba/F3 cells. (a) Venn diagram summary of results from experimental workflow after mapping of all phospho-proteomic data across all experiments. (b and c) Distribution of quantitative phosphorylation changes of tofacitinib/control and ruxolitinib/control. (d and e) Tofacitinb (500 nM) treatment resulted in 84 down- and 29 upregulated phosphorylation sites (significance reported for changes versus control of ±0.5 log2 fold). (e) Ruxolitinib (500 nM) treatment resulted in 86 down- and 20 upregulated phosphorylation sites (significance reported for changes versus control of ±0.5 log2 fold). (f) STAT5A showed consistent downregulation across all three replicates upon tofacitinib and ruxolitinib treatment in the phospho-proteomic screen. (g) Western blot analysis further validated the decrease in phospho-STAT5 (Y694) upon treatment with JAK selective inhibitors tofacitinib and ruxolitinib.
Using a cutoff of log2 fold change >0.5 we identified 84 downregulated unique phosphorylation sites and 29 upregulated phosphorylation sites following 90 min of JAK3 inhibition using tofacitinib across biological and technical replicates (Figure 2d). Similarly, treatment with ruxolitinib identified 86 downregulated and 20 upregulated phosphorylation sites (Figure 2e). In both instances, the phosphorylation of STAT5A at tyrosine Y694 was significantly reduced by −5.9- and −3.5-fold following tofacitinib or ruxolitinib treatment, respectively (Figure 2f), which was also confirmed by western blotting (Figure 2g).
The spectral intensity for each unique phosphosite (reporter-ion) within each phosphoprotein was averaged across biological and technical replicates for each treatment and grouped. The global phosphopeptide profile of JAK3 and JAK1 inhibition were clustered into canonical and biological pathways (as described in Materials and Methods, Figure 3). As expected, canonical Ingenuity Pathway Analysis pathways associated with decreased phosphosite abundance after both tofacitinib and ruxolitinib treatments was linked to JAK/STAT signaling and also IL-22 signaling. This cluster was also significantly enriched for proteins implicated in 14-3-3-mediated signaling (Figure 3, Cluster 2). Cluster 3 showed increased phosphosite abundance after tofacitinib and ruxolitinib treatments with a significant enrichment for proteins implicated DNA methylation, transcriptional repression and ERK/MAPK signaling pathways.
Identification of five independent phosphosite clusters following tofacitinib and ruxolitinib treatment. Heatmap of the average of reporter ion quantifications for each phosphosite in three replicate experiments are shown. Biological functions and signaling pathways identified by Ingenuity Pathway Analysis (IPA) have been assigned to each cluster if significantly over-represented by phosphopeptides. Fisher’s exact test was used to evaluate significance, which was set at P-values <0.01.
Of interest, cluster 1 and cluster 5 are associated with pharmacological inhibition of JAK3 specifically. These clusters are enriched for pathways involved in altered DNA Repair mediated by the homologous recombination repair pathways, multiple components of complementary apoptotic cascades such as the cytolytic granzyme B pathway, the growth arrest and DNA damage-inducible 45 (GADD45) pathway (Figure 3, Cluster 1) and also PI3K/AKT signaling and protein ubiquitination (Figure 3, Cluster 5). These data show that specific inhibition of JAK1 or JAK3 may have slightly different effects on downstream signaling pathway.
The global phosphopeptide list was then filtered to generate a short list of significantly altered changes common to both tofacitinib or ruxolitinib treatment (average ±0.5 log2-fold change/dimethylsulfoxide control) (Supplementary Table 3). This stringent short list consisted of 84 and 48 unique phosphosites decreasing or increasing in phosphorylation respectively after both tofacitinib and ruxolitinib treatment. A heatmap showing individual replicate values of phosphosite abundances compared to control is presented in Figure 4a for each protein identified. Within this list, four proteins had altered tyrosine phosphorylation, STAT5(Y694) and ACTB(Y198) sites were decreased after inhibitor treatment whilst PGRMC2(Y204) and MCM2(Y137) were increased after inhibitor treatment. The remaining changes occurred on serine and threonine residues. These proteins and their major cellular functions are illustrated schematically in Figure 4b.
Heatmap and schematic representation of phosphosites down- or upregulated common to both tofacitinib and ruxolitinib treatment. Phosphosites with an average decrease or increase of 0.5 log2 fold relative to dimethylsulfoxide (DMSO) control are shown. (a) Color reflects fold change in phosphorylation relative to control (DMSO treated) cells. Blue and yellow indicate down- and upregulation respectively. An asterisk indicates phosphosites, which could not be annotated with 100% certainty. (b) Schematic for a selection of phosphosites with an average decrease or increase of 0.5 log2 fold relative to DMSO control are shown alongside the main cellular process they are involved. Yellow and blue indicates an increase or decrease in phosphorylation, respectively upon JAK selective inhibition (tofacitinib and ruxolitinib). (c) PRM validation of phosphoproteomic profiling of JAK3(L857Q) signaling following tofacitinib or ruxolitinib treatment. (d) PRM correlation between JAK3(L857Q) and JAK3(M511) signaling following tofacitinib and ruxolitinib treatment. (e) PRM validation of phosphorylation upon JAK selective inhibition (tofacitinib and ruxolitinib) in BaF/3 JAK3(L857Q) and JAK3(M511I) transformed cells and in patient samples 389E and XC65. (PRM phosphosite annotation reflects mouse residues), n/f denotes phosphopeptide not found.
Proteins involved in RNA processing and chromatin remodeling are directly regulated downstream of mutant JAK3 signaling
There were 18 proteins associated with RNA metabolism including mRNA stability/processing, splicing and degradation some of which have established roles in cancer (Figure 4b). De novo phosphorylation of Threonine 7 (T7) of SF3B1 (splicing factor 3b, subunit 1) increased upon inhibition of JAK3 mutant signaling. This factor is important for anchoring the spliceosome to precursor mRNA and is one of the most frequently mutated genes in chronic lymphocytic leukemia.18, 19, 20 Serine 100 of RNMT showed increased phosphorylation and this protein is important in the coordination of mRNA capping and linked to cancer.21 In addition to RNA metabolism, an unexpected link with several proteins involved in epigenetic regulation was also found. HIST1H1E, a regulator of higher-order chromatin structure that is mutated in lymphoma,22 showed an increased phosphorylation on serine 36 upon JAK inhibition. MCM8, which is linked with the development of chronic myelogenous leukemia, showed a decrease in phosphorylation upon JAK inhibition.23
Confirmation of novel phosphorylation changes in proteins regulating RNA processing and chromatin remodeling was performed by an independent targeted proteomics experiment (parallel reaction monitoring, PRM) (Figures 4c–e and Supplementary Table 2). This approach revealed a significant level of correlation between methods of phosphopeptide quantification (that is, discovery by data-dependent acquisition and validation by PRM) and also between JAK3(L867Q) and JAK3(M511I) mutants (Figures 4c and d). Reduction in relative fold-change is a consequence of data-dependent acquisition based quantitative approaches (reporter-ion based relative quantification—TMT Figure 4a), owing to the thousands of events measured across nine samples simultaneously.24 Therefore, changes in phosphorylation in proteins such as STAT5 and YBX3 were more accurately measured using targeted methods such as PRM (Figure 4e). Nevertheless, discovery proteomics is a powerful tool that guided our PRM validation study and revealed a highly significant correlation between phosphoproteins regulating RNA processing and chromatin remodeling in both JAK3 L857Q- and M511I-transformed cells.
Human JAK3 mutant T-ALL samples 389E and XC65 (Supplementary Table 1) were also validated by PRM. Significant correlation of the abundance of phosphoproteins regulated by mutant JAK3 were seen between patients following JAK1 inhibition using ruxolitinib (Figure 4e and Supplementary Figure 1A). STAT5 and proteins implicated in DNA methylation (DNMT1 and KDM2A), genome replication (MCM8 and NCAPD2) and pre-mRNA processing (SF1 and NCBP1) were significantly modulated in both patient samples following JAK inhibition (Figure 4e and Supplementary Table 2). However, regulation of phosphoproteins following mutant JAK3 inhibition using tofacitinib was not as consistent as with ruxolitinib, indicating again differences between these drugs (Figure 4e).
To further complement the PRM, western blot analysis was used to confirm the phosphorylation status for EIF4EBP1 at threonine T45 and RANBP3 at serine S58, for which site-specific phospho-antibodies were available. Both showed significantly reduced phosphorylation following treatment with tofacitinib or ruxolitinib (Supplementary Figure S1B). Moreover, these phosphorylation sites were confirmed as being directly downstream of JAK3 (L857Q and M511I) mutations with Ba/F3 transformed by the activating mutation STAT5 N642H or parental Ba/F3 stimulated with IL3 showed significantly less phosphorylation at these sites compared with untreated Ba/F3 cells expressing either of these JAK3 mutations (Supplementary Figure S1B).
Inhibiting JAK in combination with the MAPK, PI3K and Bcl-2 pathway results in synergistic or additive effects on Ba/F3 cells transformed by JAK3 mutants
Although a large number of novel phosphorylation changes were identified downstream of mutant JAK3 signaling including in RNA metabolism and chromatin remodeling, the majority of these proteins remain difficult to target due to the lack of small molecular inhibitors. To this end, pathway interrogation using Ingenuity Pathway Analysis revealed multiple components of MAPK, PI3K and apoptosis pathway to be modulated following either tofacitinib or ruxolitinib treatment (Figure 5a, Supplementary Figures S2–S4 and Supplementary Table S4). Specifically, phosphorylation changes were seen in SHC, GRB2 and GAB1/2, which are upstream of the MEK/ERK pathway. Moreover, GAB1/2 is also known to associate with the PI3K pathway (Figure 5b). In line with this analysis, acute inhibition of JAK3(L857Q)- and JAK3(M511I)-transformed Ba/F3 cells with tofacitinib revealed significantly decreased pERK1/2 and pMEK1 levels. This was further reduced when given in combination with the MEK inhibitors selumetinib and trametinib (Figure 5c). Although the phosphoproteomics data did not identify phosphorylation changes in any anti- or pro-apoptotic proteins, mutant JAK3 signaling would be expected to upregulate members of the anti-apoptotic BCL2 family including BCL-2 and MCL1 at the transcriptional level.25 Analysis of expression data from 264 T-ALL patients26 revealed that cases with JAK3 mutations (n=20) had significantly higher levels of BCL2 compared with non-mutated cases. There were no differences for BCL-XL or MCL1 expression (Figure 5d).
Ingenuity Pathway Analysis (IPA) identifies significant enrichment for PI3K/AKT, MEK/ERK and apoptosis signaling. (a) Pathways for which phospho-sites showed a 0.5 log2 fold change increase or decrease in phosphorylation compared to control following tofacitinib or ruxolitinib treatment are shown. (b) Schematic representation of PI3K, apoptosis, and MEK/ERK pathway. Proteins that show an increase or decrease in phosphorylation by tofacitinib or ruxolitinib treatment (average ±0.5 log2 fold change /dimethylsulfoxide (DMSO) control) are indicated with an asterisk. Inhibitors used in downstream experiments are indicated. (c) Western blot assessment of downstream STAT5, ERK and MEK activation in JAK3(L857Q) or JAK3(M511I) transformed Ba/F3 cells treated with tofacitinib, selumtinib or trametinib for 90 min. (d) RNA-seq analysis of BCL2, MCL1 and BCL-XL in T-ALL patients that either have a JAK3 mutation (n=20), other JAK-STAT pathway mutations (n=45, includes mutations in IL7R, PTPN2, FLT3, STAT5B, JAK1, JAK3, SH2B3, IL2RB) or no mutations in JAK-STAT (n=199). (P-value calculated using welch t-test).
These data therefore provided the rationale to test whether selective inhibitors of these pathways such as the selective Bcl-2 inhibitor (ABT-199), PI3K inhibitor (buparlisib) or MEK inhibitors (selmuetinib and trametinib) would synergistically inhibit cellular growth and survival when used in combination with a JAK selective inhibitor.
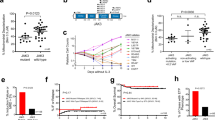
Ba/F3 cells transformed by two different JAK3 mutants, JAK3(M511I) or JAK3(L857Q), were treated with a combination of various concentrations of tofacitinib and the MEK inhibitor selumetinib. Most combinations showed a synergistic effect ranging from moderate (0.3–0.05 μM tofacitinib and 0.89–1.582 μM selumetinib) to high synergism (0.3 μM tofacitinib and 2.109–5 μM selumetinib) (Figure 6a). Interestingly, all combinations of tofacitinib with the MEK inhibitor trametinib showed synergistic cytotoxicity (CI <0.1–0.9) in a dose-dependent manner (Figure 6a). This combination had no effect on parental Ba/F3 cells stimulated by IL3 (data not shown).
Inhibiting JAK in combination with the MAPK, PI3K and Bcl-2 pathway results in synergistic or additive effects on Ba/F3 cells transformed by JAK3 mutants. (a) Chou-Talalay plots showing the effect of tofacitinib with MEK inhibitors (selumetinib, trametinib), Bcl-2 inhibitor (ABT-199) or PI3K inhibitor (buparlisib) on Ba/F3 cells transformed by JAK3 M511I or L857Q after 24 h incubation. CompuSyn was used to calculate the combination index (CI). CI<1 indicate synergistic effects, C=1 indicate additive effects, C>1 indicate antagonistic effects. (b) Ba/F3 transformed by M511I or L857Q were treated for 24 h with single compounds or combination. Relative proliferation compared to vehicle treated cells is shown. Data represents the average of three experiments±s.e.m. Significance was calculated using One-way analysis of variance (ANVA) and the Bonferroni correction. (****P⩽0.0001; ***P⩽0.001; **P⩽0.01). Concentrations used; 0.15 μM tofacitinib in combination with 3.75 μM selumetinib, 0.67 μM trametinib, 0.6 μM buparlisib and 0.3 μM tofacitinib in combination with 0.15 μM ABT-199.
Inhibition of the anti-apoptotic protein Bcl-2 using ABT-199 in combination with tofacitinib was also synergistic at moderate concentrations. However, low concentrations of both tofacitinib and ABT-199 were antagonistic, with CI values reaching 2.7. Effects from intermediate concentration combinations vary from moderate synergism to highly synergistic. The combination of 0.3 μM ABT-199 and 0.3 μM tofacitinib induced synergistic cytotoxicity (CI values up to 0.3).
Inhibition of the P13K pathway revealed both high synergism and high antagonism dependent on dosing schedules. Low dosage of tofacitinib (up to 0.05 μM) in combination with buparlisib elicited a highly antagonistic effect. However, significant synergism (CI<0.1) was elicited through the use of intermediate tofacitinib concentrations (0.2 μM) in combination with high dose buparlisib (2.5 μM).
These initial screening matrices were then used to determine optimal dosage regimens required to elicit a synergistic cytotoxicity in mutant JAK3-expressing cells and subsequently to be used in experiments at fixed concentrations, alone and combined. These fixed dose regimens significantly reduced proliferation when in combination compared with single compound treatments for both JAK3(M511I) and JAK3(L857Q) mutant-transformed Ba/F3 cells (Figure 6b).
Synergistic effect of tofacitinib in combination with MEK or Bcl-2 inhibitors in JAK3 mutant PDX samples
To determine the preclinical efficacy of these drug combinations, we next sought to determine the cytotoxic effect on primary patient-derived T-ALL cells. Two T-ALL samples expressing a JAK3(M511I) mutation (X1 1and 389E) and one sample that was JAK3 wild type (X10) were used (Supplementary Table 1). These patient samples were initially expanded within NSG mice before collection of the leukemic cells and immediately incubated ex vivo with our optimized JAK3 pathway targeting regimens for 48 h (Figure 7a).
Synergistic effect of tofacitinib in combination with MEK or Bcl-2 inhibitors in JAK3 mutant PDX samples. (a) Schematic representation of experimental workflow. (b) Chou-Talalay plots showing the effect of tofacitinib with MEK inhibitors (selumetinib, trametinib) or Bcl-2 inhibitor (ABT-199) on T-ALL-derived PDX samples. CompuSyn was used to calculate the combination index (CI). CI<1 indicate synergistic effects, C=1 indicate additive effects, C>1 indicate antagonistic effects. (c–e) Relative viable cell count is after 48 h treatment of PDX samples with a combination of 0.2 μM tofacitinib and 0.3 μM selumetinib (c) or 0.2 μM trametinib (d) or a combination with 0.3 μM tofacitinib and 0.13 μM ABT-199 (e). Data represents the average of three experiments±s.e.m. Significance was calculated using one-way analysis of variance (ANOVA) and the Bonferroni correction. (****P⩽0.0001; ***P⩽0.001; nonsignificant (ns) P⩾0.05).
Tofacitinib in combination with the MEK and Bcl-2 inhibitors were selected, as these combinations showed the most consistent synergic effects over a wide range of concentrations. Synergy was initially assessed and confirmed using the T-ALL X11 sample (JAK3 M511I) for the combination of tofacitinib and the MEK inhibitors, selumetinib and trametinib, and the Bcl-2 inhibitor ABT-199. Although most concentration combinations showed synergic efficacy, some combinations of selumetinib and tofacitinib were antagonistic revealing CI values up to 1.7. Notably, this antagonism was only seen when combinations had the highest concentration of tofacitinib (1.28 μM) (Figure 7b). Importantly, the combination of trametinib and tofacitinib elicited cytotoxicity with CI values ranging from <0.1 to 1, which is indicative of highly synergistic efficacy. Furthermore, the most synergic combination was elicited through the combination of low-dose tofacitinib (0.01–0.04 μM) with low-dose trametinib concentrations (0.015 μM) (Figure 7b). Targeting BCL2 using ABT-199 in combination with tofacitinib revealed high synergism. Low-dose tofacitinib (0.08 μM) with low-dose ABT-199 (0.044 μM) resulted in a highly synergistic CI value of <0.1 (Figure 7b).
Single synergistic dosage combinations determined using the X11 sample were then used on the 389E (JAK3 M511I mutant T-ALL sample) and X10 (JAK3 Wild-Type T-ALL sample) samples (Figures 7c and d). The T-ALL cells expressing a JAK3(M511I) mutation showed significantly reduced viability upon treatment with the combination of the JAK selective inhibitor tofacitinib and a MEK inhibitor selumetinib (Figure 7c) or trametinib (Figure 7d) compared with single-compound treatment. Interestingly, T-ALL cells 389E showed less sensitivity to JAK selective inhibition alone (compared with X11), but synergy was still observed. The T-ALL sample × 10, which expresses wild-type JAK3 (and also does not harbor IL7R or JAK1 mutations), did not show any reduction in cell viability after treatment with tofacitinib and/or selumetinib and trametinib showing that the synergy is dependent on the presence of a JAK3 mutation (Figures 7c and d).
The T-ALL samples were also treated with the Bcl-2 inhibitor ABT-199 alone or in combination with tofacitinib. T-ALL samples with JAK3 mutation were sensitive to both tofacitinib or ABT-199 alone, and the combination of these drugs was more effective than each drug separately. The T-ALL sample × 10 (JAK3 wild type) showed reduced cell viability after treatment with the Bcl-2 inhibitor ABT-199, but there was no additional reduction when combined with the JAK-selective inhibitor tofacitinib (Figure 7e), indicating again that synergy between tofacitinib and ABT-199 is only obtained in T-ALL samples harboring a JAK3 mutation. Similar results were shown after apoptosis staining using Annexin-V and propidium iodide. Treatment of the T-ALL sample X11 with tofacitinib in combination with selumetinib, trametinib or ABT-199 resulted in an increase in Annexin-V+/propidium iodide+ cells compared with cells treated with a single compound (Supplementary Figure S5).
In vivo combination treatment of patient-derived xenograft sample carrying a JAK3 mutations leads to a significant decrease in leukemia burden
Having established strong synergism between JAK3 and Bcl-2 inhibition, we moved to determine whether this could also be observed in vivo. To this end, an in vivo patient-derived xenograft model was established with T-ALL sample 389E that carries a JAK3 (M511I) mutation. For these studies, the JAK1-selective inhibitor ruxolitinib was used due to its known side effect profile and known efficacy in other hematological disorders27 in combination with ABT-199. To accurately detect synergism, suboptimal concentrations were used for ABT-199 (20 mg/kg/day/PO) and ruxolitinib (40 mg/kg/day/PO), which were 70–80% reduced compared with previously published studies when used as a single agent.28, 29 Leukemia burden was assessed by human CD45 expression in the peripheral blood at the beginning, midpoint and end stage of the 14-day treatment phase. After 14 days, disease burden was assessed in all mice and the combination treatment of ruxolitinib and ABT-199 led to significant decrease of leukemia cells in the peripheral blood and significantly less bone marrow compared with vehicle, ABT-199-only or ruxolitinib-only treatment (Figures 8a–c).
Combination treatment of mice xenografted with JAK3 (M511I) patient 389E. (a) Leukemia burden was followed on basis of human CD45 expression in the peripheral blood. Treatment with vehicle (n=7), ruxolitinib (40 mg/kg/day)(n=7), ABT-199 (20 mg/kg/day; n=7) or ruxolitinib (40 mg/kg/day)+ABT-199 (20 mg/kg/day) (n=8) was begun once CD45%>2% and indicated with an arrow. Treatment continued daily for 14 days. (b) End-stage assessment of human (h)CD45% in the peripheral blood. (c) End-stage assessment of human (h)CD45% in the bone marrow. Significance was calculated using student t-test (***P⩽0.01; **P⩽0.05).
Discussion
Although improved treatment and supportive care have led to a better outcome for T-ALL patients, poor response to therapy and relapse remain significant problems, especially in adult patients. In addition, survivors continue to suffer long-term effects from chemotherapy and in adults the rates of survival remain below 50%.30, 31 Although there is no evidence that JAK3 mutations are associated with a poor prognosis, this tyrosine kinase is an attractive therapeutic target to help establish a more patient-specific therapy.2, 9 Precision therapeutic strategies targeting recurring oncogene addicted cells along with the pathways that cooperate in malignant cell growth offer treatment potentials that are believed to have the greatest efficacy, while reducing side effects.
Here we describe the first comprehensive analysis of phosphoproteome signaling pathways regulated downstream of mutant JAK3. We identified changes in phospho-peptides upon inhibition of mutant JAK3 signaling, with the identification of proteins involved in MAPK, PI3K/mTOR and apoptosis, and hitherto unknown links to proteins involved RNA processing and epigenetic regulation. It is known that JAK proteins are able to phosphorylate the SH2 domain containing protein SHC1 that is recruited to the cytokine receptor and in turn recruits the GRB2–SOS complex. SOS is the guanosine nucleotide exchange factor for RAS. Through RAS, the GRB2-SOS complex is able to activate the canonical MAPK pathway.32 Both SHC1 and GAB2 phosphorylation were significantly decreased upon inhibition of mutant JAK3 signaling. In addition to the RAS/MAPK pathway, the PI3K pathway has also been shown to have an important role downstream of the JAK kinases. Sharfe et al.5 showed that JAK3 was able to interact with the p85 subunit of PI3K upon IL7 stimulation and subsequently induced phosphorylation of this p85 subunit. PI3K also interacts with the GAB2 adaptor protein, which indicates a link between PI3K and MAPK activation.33
A number of candidate proteins found to be directly downstream of mutant JAK3 have already been found to have important roles either in the pathogenesis of T-ALL or cancer in general. We have recently described a link between mutations in proteins involved in epigenetic regulation such as the PRC2 complex and mutations in the IL7R/JAK3 signaling pathway.2 Here we show phosphoproteomic data linking mutant JAK3 signaling with proteins implicated in epigenetic regulation. In the current work, we describe an increased phosphorylation of KDM2A upon inhibition of mutant JAK3 signaling. KDM2A is a histone demethylase that specifically demethylates lysine-36 of histone 3. Increased expression levels of KDM2A promotes tumor cell growth and migration in gastric cancer34 but there are currently no reports on how the phosphorylation status of KDM2A threonine 713 (T713) regulates its activity. However, KDM2A is phosphorylated at threonine 632 (T632) following DNA damage and largely abrogates the chromatin-binding capacity of KDM2A.35 We also reveal reduced phosphorylation of the DNA methyltransferase family member DNMT1 upon inhibition of mutant JAK3 signaling (Figure 4). DNMT1 is required to maintain the methylation of the entire genome and altered expression has been observed in a range of cancers including lymphoma, breast, colon, liver, pancreas and esophagus cancer.36 For both KDM2A and DNMT1, it will be important now to determine the role of these phosphorylation sites in regulating protein function and their role in T-ALL.
One of the more surprising links we found downstream mutant JAK3 signaling was RNA metabolism including proteins involved in mRNA stability, splicing and degradation. There were significant decreased phosphorylation changes upon inhibition of JAK3 in RRP1B, NCBP1, SERBP1, YBX3, ATXN2L, U2SURP, HNRNPM, HNRNPK and GTPBP1, and significant increased phosphorylation changes upon inhibition of JAK3 in CMTR1, RNMT, ZC3H13, SF3B1, PNN, CASC3 and SF1. Of these candidate proteins, a number already have potential roles in cancer. RRP1B expression levels can predict patient outcome in breast cancer and can regulate histone methylation.37 SERBP1 has been found to be significantly upregulated in CD34+ CML cases compared with control C34+ cells.38 Significant hypermethylation of the YBX3 promoter to downregulate its mRNA expression has been found in AML.39 HNRNPM has been found to promote cancer metastasis by regulating alternative splicing.40 Depletion of RNMT has been described to effectively and specifically inhibits cancer cell growth and cell invasive capacities in different types of cancer.21 SRRM1 has been shown to regulate splicing of the CD44 protein41 and the identification of different residues that increase and decrease in phosphorylation status may indicate a functional switch for this protein. Finally, SF3B1 is recurrently mutated in MDS and CLL,20, 42 and SF3B1 inhibitors are in preclinical development for the treatment of cancer and therefore may have potential indications in combination with JAK3 inhibitors in T-ALL.
To this end, phosphoproteomic profile provided a list of pathways that could be targeted by small molecular inhibitors in combination of JAK3 inhibitors. We show that the combined inhibition of JAK3 signaling with inhibitors targeting PI3K, MEK or BCL2 resulted in a synergistic cytotoxic effect in Ba/F3 samples transformed by JAK3 mutants and primary T-ALL patient samples carrying JAK3 mutations. This synergistic effect was selective for JAK3-mutated T-ALL samples, as no synergy was observed in the T-ALL sample that was wild type for JAK3. We also tested the combination of ruxolitinib with ABT-199 in vivo and this also resulted in a significant decrease in leukemia burden, suggesting this combinational would be an effective treatment strategy in patients with a JAk3 mutation. Recently, Canté-Barrett et al.43 showed that inhibiting both the MEK and PI3K-AKT pathways synergistically prevents the proliferation of Ba/F3 cells expressing JAK3 mutants. Furthermore, combined inhibition of MEK and PI3K/AKT was cytotoxic to four primary T-ALL patient samples, which carried a JAK3 mutation. It has also been shown that immature T-ALL patients with high levels of BCL2 were sensitive to ABT-199 as a single agent.44 We have now extended this observation to take into account the JAK3 mutational status and tested the inhibitory effects of ABT-199 in combination with tofacitinib, a JAK selective inhibitor and show that this combination is highly effective in causing cytotoxicity for T-ALL cells. It is important to note that for some concentrations combinations of MEK or BCL2 inhibitors with the JAK inhibitor tofacitinib resulted in an antagonistic or additive effect rather than a synergistic effect. This highlights the importance of identifying the optimal treatment dose for patients in order to obtain maximal synergy.
In summary, this study has characterized signaling downstream of mutant JAK3 showing constitutive activation of proteins involved in cell cycle, translation, apoptosis, MAPK and PI3K/AKT pathways and epigenetic regulation. Through the use of primary T-ALL samples we show synergistic inhibition by combining JAK-selective inhibitors with MEK, PI3K and BCL2 inhibitors. Together, our research gives the incentive to further explore the use of JAK inhibitors in combination with MEK or BCL2 inhibitors in order to optimize current treatment regimes.
Change history
19 September 2018
Following the publication of this article the authors noted that data describing precisely where phosphorylation sites in proteins modulated following JAK1 or JAK3 inhibition in mutant T-ALL samples was not clearly annotated. Therefore an additional sheet has been added to Supplementary Table 2. The authors wish to apologize for any inconvenience caused.
References
Stock W, La M, Sanford B, Bloomfield CD, Vardiman JW, Gaynon P et al. What determines the outcomes for adolescents and young adults with acute lymphoblastic leukemia treated on cooperative group protocols? A comparison of Children's Cancer Group and Cancer and Leukemia Group B studies. Blood 2008; 112: 1646–1654.
Vicente C, Schwab C, Broux M, Geerdens E, Degryse S, Demeyer S et al. Targeted sequencing identifies association between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica 2015; 100: 1301–1310.
Degryse S, de Bock CE, Cox L, Demeyer S, Gielen O, Mentens N et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood 2014; 124: 3092–3100.
Osinalde N, Sanchez-Quiles V, Akimov V, Guerra B, Blagoev B, Kratchmarova I . Simultaneous dissection and comparison of IL-2 and IL-15 signaling pathways by global quantitative phosphoproteomics. Proteomics 2015; 15: 520–531.
Sharfe N, Dadi HK, Roifman CM . JAK3 protein tyrosine kinase mediates interleukin-7-induced activation of phosphatidylinositol-3' kinase. Blood 1995; 86: 2077–2085.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Girardi T, Vicente C, Cools J, De Keersmaecker K . The genetics and molecular biology of T-ALL. Blood 2017; 129: 1113–1123.
De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 2013; 45: 186–190.
Degryse S, Cools J . JAK kinase inhibitors for the treatment of acute lymphoblastic leukemia. J Hematol Oncol 2015; 8: 91.
Losdyck E, Hornakova T, Springuel L, Degryse S, Gielen O, Cools J et al. Distinct acute lymphoblastic leukemia (ALL)-associated janus kinase 3 (JAK3) mutants exhibit different cytokine-receptor requirements and JAK inhibitor specificities. J Biol Chem 2015; 290: 29022–29034.
Dun MD, Chalkley RJ, Faulkner S, Keene S, Avery-Kiejda KA, Scott RJ et al. Proteotranscriptomic profiling of 231-BR breast cancer cells: identification of potential biomarkers and therapeutic targets for brain metastasis. Mol Cell Proteomics 2015; 14: 2316–2330.
Fujiki Y, Hubbard AL, Fowler S, Lazarow PB . Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 1982; 93: 97–102.
Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 2003; 75: 1895–1904.
Engholm-Keller K, Birck P, Størling J, Pociot F, Mandrup-Poulsen T, Larsen MR . TiSH—a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J Proteomics 2012; 75: 5749–5761.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010; 26: 966–968.
Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Dun MD et al. Next generation sequencing analysis reveals segmental patterns of microRNA expression in mouse epididymal epithelial cells. PLoS One 2015; 10: e0135605.
Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 2011; 10: 5354–5362.
Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2011; 44: 47–52.
Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506.
Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 2011; 118: 6904–6908.
Stefanska B, Cheishvili D, Suderman M, Arakelian A, Huang J, Hallett M et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res 2014; 20: 3118–3132.
Lunning MA, Green MR . Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J 2015; 5: e361.
Cai L, Zhao K, Yuan X . Expression of minichromosome maintenance 8 in chronic myelogenous leukemia. Int J Clin Exp Pathol 2015; 8: 14180–14188.
Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS . Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics 2010; 9: 1885–1897.
Takada K, Jameson SC . Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol 2009; 9: 823–832.
Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017; 63: 5329.
Senkevitch E, Durum S . The promise of Janus kinase inhibitors in the treatment of hematological malignancies. Cytokine 2016; 98: 33–41.
Chonghaile TN, Roderick JE, Glenfield C, Ryan J, Sallan SE, Silverman LB et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov 2014; 4: 1074–1087.
Evrot E, Ebel N, Romanet V, Roelli C, Andraos R, Qian Z et al. JAK1/2 and Pan-deacetylase inhibitor combination therapy yields improved efficacy in preclinical mouse models of JAK2V617F-driven disease. Clin Cancer Res 2013; 19: 6230–6241.
Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. J Clin Oncol 2012; 30: 1663–1669.
DeAngelo DJ, Stevenson KE, Dahlberg SE, Silverman LB, Couban S, Supko JG et al. Long-term outcome of a pediatric-inspired regimen used for adults aged 18-50 years with newly diagnosed acute lymphoblastic leukemia. Leukemia 2015; 29: 526–534.
Osinalde N, Moss H, Arrizabalaga O, Omaetxebarria MJ, Blagoev B, Zubiaga AM et al. Interleukin-2 signaling pathway analysis by quantitative phosphoproteomics. J Proteomics 2011; 75: 177–191.
Gadina M, Sudarshan C, Visconti R, Zhou YJ, Gu H, Neel BG et al. The docking molecule gab2 is induced by lymphocyte activation and is involved in signaling by interleukin-2 and interleukin-15 but not other common gamma chain-using cytokines. J Biol Chem 2000; 275: 26959–26966.
Huang Y, Liu Y, Yu L, Chen J, Hou J, Cui L et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol 2015; 36: 271–278.
Cao L-L, Wei F, Du Y, Song B, Wang D, Shen C et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 2016; 35: 301–313.
Zhang W, Xu J . DNA methyltransferases and their roles in tumorigenesis. Biomark Res 2017; 5: 1.
Lee M, Dworkin AM, Lichtenberg J, Patel SJ, Trivedi NS, Gildea D et al. Metastasis-associated protein ribosomal RNA processing 1 homolog B (RRP1B) modulates metastasis through regulation of histone methylation. Mol Cancer Res 2014; 12: 1818–1828.
Čokić VP, Mojsilović S, Jauković A, Kraguljac-Kurtović N, Mojsilović S, Šefer D et al. Gene expression profile of circulating CD34(+) cells and granulocytes in chronic myeloid leukemia. Blood Cells Mol Dis 2015; 55: 373–381.
Wong JJ-L, Lau KA, Pinello N, Rasko JEJ . Epigenetic modifications of splicing factor genes in myelodysplastic syndromes and acute myeloid leukemia. Cancer Sci 2014; 105: 1457–1463.
Xu Y, Gao XD, Lee J-H, Huang H, Tan H, Ahn J et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 2014; 28: 1191–1203.
Cheng C, Sharp PA . Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol 2006; 26: 362–370.
Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jädersten M, Jansson M et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015; 126: 233–241.
Canté-Barrett K, Spijkers-Hagelstein JAP, Buijs-Gladdines JGCAM, Uitdehaag JCM, Smits WK, van der Zwet J et al. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016; 30: 1832–1843.
Peirs S, Matthijssens F, Goossens S, Van de Walle I, Ruggero K, de Bock CE et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood 2014; 124: 3738–3747.
Acknowledgements
Dr Ben Crossett from the Mass Spectrometry Core Facility at The University of Sydney University, Mr Nathan Smith from The University of Newcastle Analytical and Biomolecular Research Facility (ABRF) and Dr Michael Mariani from Thermo Fisher Scientific provided MS support. The Academic and Research Computing Support (ARCS) team, within IT Services at the University of Newcastle, provided high performance computing (HPC) infrastructure for supporting the bioinformatics. This study was supported by grants from the Belgian government (cancer plan), the FWO-Vlaanderen (JC and PVV), the Children Cancer Fund Ghent (PVV), the Belgian Stand Up To Cancer Foundation (PVV) and the Foundation Against Cancer (JC and PVV); a European Research Council grant (JC); the Interuniversity Attraction Poles granted by the Federal Office for Scientific, Technical and Cultural Affairs, Brussels, Belgium (JC); and the Agency for Innovation by Science and Technology in Flanders, Belgium (SD and DB); Cancer Institute NSW, Australia ECF (MDD and NV). The Hunter Cancer Research Alliance and the European Association for Cancer Research provided a travel scholarship for S Degryse. IG and SB are both Aspirant Fellows of the FWO-Vlaanderen. Jennie Thomas, Life Governor of the Hunter Medical Research Institute provided a travel scholarship for MDD. The Cancer Institute NSW in partnership with the Faculty of Health and Medicine from the University of Newcastle funded the MS platform.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
SB is an employee of Thermo Fisher Scientific, the corporation that produces Orbitrap mass spectrometers and proteomics software. SB is an LCMS applications specialist and provided technical support for MS methodology and data interpretation. Beyond this, the authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this manuscript. There are no patents, products in development, or marketed products to declare. All other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Degryse, S., de Bock, C., Demeyer, S. et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 32, 788–800 (2018). https://doi.org/10.1038/leu.2017.276
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.276
This article is cited by
-
Targeted therapy and immunotherapy for T cell acute lymphoblastic leukemia/lymphoma
Annals of Hematology (2023)
-
JAK3 mutations and mitochondrial apoptosis resistance in T-cell acute lymphoblastic leukemia
Leukemia (2022)
-
Phosphoproteomic profiling of T cell acute lymphoblastic leukemia reveals targetable kinases and combination treatment strategies
Nature Communications (2022)
-
Phospho-heavy-labeled-spiketide FAIMS stepped-CV DDA (pHASED) provides real-time phosphoproteomics data to aid in cancer drug selection
Clinical Proteomics (2022)
-
A multiparametric niche-like drug screening platform in acute myeloid leukemia
Blood Cancer Journal (2022)