Abstract
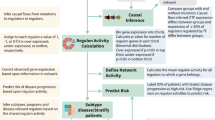
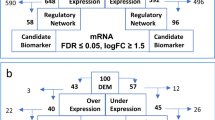
Multiple myeloma (MM) is an incurable malignancy of bone marrow plasma cells characterized by wide clinical and molecular heterogeneity. In this study we applied an integrative network biology approach to molecular and clinical data measured from 450 patients with newly diagnosed MM from the MMRF (Multiple Myeloma Research Foundation) CoMMpass study. A novel network model of myeloma (MMNet) was constructed, revealing complex molecular disease patterns and novel associations between clinical traits and genomic markers. Genomic alterations and groups of coexpressed genes correlate with disease stage, tumor clonality and early progression. We validated CDC42BPA and CLEC11A as novel regulators and candidate therapeutic targets of MMSET-related myeloma. We then used MMNet to discover novel genes associated with high-risk myeloma and identified a novel four-gene prognostic signature. We identified new patient classes defined by network features and enriched for clinically relevant genetic events, pathways and deregulated genes. Finally, we demonstrated the ability of deep sequencing techniques to detect relevant structural rearrangements, providing evidence that encourages wider use of such technologies in clinical practice. An integrative network analysis of CoMMpass data identified new insights into multiple myeloma disease biology and provided improved molecular features for diagnosing and stratifying patients, as well as additional molecular targets for therapeutic alternatives.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Röllig C, Knop S, Bornhäuser M . Multiple myeloma. Lancet 2015; 385: 2197–2208.
Siegel RL, Miller KD, Jemal A . Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Szalat R, Munshi NC . Genomic heterogeneity in multiple myeloma. Curr Opin Genet Dev 2015; 30: 56–65.
Jonathan J, Keats DWC, Liang W, Venkata Y, Kurdoglu A, Aldrich J et al. Interim analysis of the MMRF CoMMpass Trial, a longitudinal study in multiple myeloma relating clinical outcomes to genomic and immunophenotypic profiles. Blood 2013; 122.
Langfelder P, Horvath S . WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014; 25: 91–101.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014; 5: 2997.
Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood 2010; 116: 2543–2553.
Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S et al. The molecular classification of multiple myeloma. Blood 2006; 108: 2020–2028.
Shaughnessy Jr JD, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 2007; 109: 2276–2284.
Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B et al. A gene expression signature for high-risk multiple myeloma. Leukemia 2012; 26: 2406–2413.
Kalff A, Spencer A . The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer J 2012; 2: e89.
Xie Z, Chng WJ . MMSET: role and therapeutic opportunities in multiple myeloma. Biomed Res Int 2014; 2014: 636514.
Mirabella F, Wu P, Wardell CP, Kaiser MF, Walker BA, Johnson DC et al. MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J 2013; 3: e114.
Xie Z, Bi C, Chooi JY, Chan ZL, Mustafa N, Chng WJ . MMSET regulates expression of IRF4 in t(4;14) myeloma and its silencing potentiates the effect of bortezomib. Leukemia 2015; 29: 2347–2354.
Mulligan G, Lichter DI, Di Bacco A, Blakemore SJ, Berger A, Koenig E et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood 2014; 123: 632–639.
Yu Y, Wang XY, Sun L, Wang YL, Wan YF, Li XQ et al. Inhibition of KIF22 suppresses cancer cell proliferation by delaying mitotic exit through upregulating CDC25C expression. Carcinogenesis 2014; 35: 1416–1425.
Prasanth SG, Prasanth KV, Stillman B . Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 2002; 297: 1026–1031.
Ishii H, Inageta T, Mimori K, Saito T, Sasaki H, Isobe M et al. Frag1, a homolog of alternative replication factor C subunits, links replication stress surveillance with apoptosis. Proc Natl Acad Sci USA 2005; 102: 9655–9660.
Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF . Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 2006; 7: 40.
Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011; 117: 211–220.
Moreaux J, Klein B, Bataille R, Descamps G, Maiga S, Hose D et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 2011; 96: 574–582.
Hiraoka A, Yano KiK, Kagami N, Takeshige K, Mio H, Anazawa H et al. Stem cell growth factor: in situ hybridization analysis on the gene expression, molecular characterization and in vitro proliferative activity of a recombinant preparation on primitive hematopoietic progenitor cells. Hematol J 2001; 2: 307–315.
Geijtenbeek TB, Gringhuis SI . Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 2009; 9: 465–479.
Yan H, Kamiya T, Suabjakyong P, Tsuji NM . Targeting C-type lectin receptors for cancer immunity. Front Immunol 2015; 6: 408.
Ingersoll SB, Ahmad S, Thoni ND, Ahmed FH, Monahan KA, Edwards JR . Targeting the IL-6 pathway in multiple myeloma and its implications in cancer-associated gene hypermethylation. Med Chem 2011; 7: 473–479.
Rosean TR, Tompkins VS, Tricot G, Holman CJ, Olivier AK, Zhan F et al. Preclinical validation of interleukin 6 as a therapeutic target in multiple myeloma. Immunol Res 2014; 59: 188–202.
Nardiello T, Jungbluth AA, Mei A, Diliberto M, Huang X, Dabrowski A et al. MAGE-A inhibits apoptosis in proliferating myeloma cells through repression of Bax and maintenance of survivin. Clin Cancer Res 2011; 17: 4309–4319.
Holien T, Misund K, Olsen OE, Baranowska KA, Buene G, Borset M et al. MYC amplifications in myeloma cell lines: correlation with MYC-inhibitor efficacy. Oncotarget 2015; 6: 22698–22705.
Holien T, Vatsveen TK, Hella H, Waage A, Sundan A . Addiction to c-MYC in multiple myeloma. Blood 2012; 120: 2450–2453.
Soodgupta D, Pan D, Cui G, Senpan A, Yang X, Lu L et al. Small molecule MYC inhibitor conjugated to integrin-targeted nanoparticles extends survival in a mouse model of disseminated multiple myeloma. Mol Cancer Ther 2015; 14: 1286–1294.
Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X . Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014; 9: e78644.
Wang Z, Gerstein M, Snyder M . RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63.
Acknowledgements
We thank the Mount Sinai Hematological Malignancies Tissue Bank (HMTB), Human Immune Monitoring Core (HIMC), Tisch Cancer Institute (TCI) (NCI Support Grant: P30 CA196521) and the Multiple Myeloma philanthropic fund for their support. This work was also supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. We also thank Katya Ahr for helping with language revision.
Author contributions
AL, JTD and SP coordinated overall study design and analysis. AL conceived and implemented the computational pipeline for data analysis. DP designed and performed laboratory experiments for hub module validation. VL, P-YK, BR and BAK performed analysis and lab experiments. DM performed clonality analysis. JK provided sequencing data. AC, HYC, BB, SJ, MDR, JY, DA and SL contributed patient samples and clinical data. JTD and SP supervised the analysis. AL, JTD and SP wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Laganà, A., Perumal, D., Melnekoff, D. et al. Integrative network analysis identifies novel drivers of pathogenesis and progression in newly diagnosed multiple myeloma. Leukemia 32, 120–130 (2018). https://doi.org/10.1038/leu.2017.197
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.197
This article is cited by
-
Aberrant non-canonical NF-κB signalling reprograms the epigenome landscape to drive oncogenic transcriptomes in multiple myeloma
Nature Communications (2024)
-
Gene interaction network analysis in multiple myeloma detects complex immune dysregulation associated with shorter survival
Blood Cancer Journal (2023)
-
Targeted single-cell proteomic analysis identifies new liquid biopsy biomarkers associated with multiple myeloma
npj Precision Oncology (2023)
-
FOXM1 regulates glycolysis and energy production in multiple myeloma
Oncogene (2022)
-
Non-small-cell lung cancer classification via RNA-Seq and histology imaging probability fusion
BMC Bioinformatics (2021)