Abstract
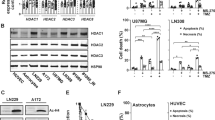
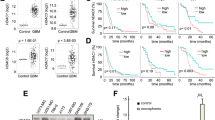
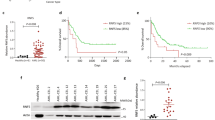
Resistance to cytotoxic chemotherapy drugs remains as the major cause of treatment failure in acute myeloid leukemia. Histone deacetylases (HDAC) are important regulators to maintain chromatin structure and control DNA damage; nevertheless, how each HDAC regulates genome stability remains unclear, especially under genome stress conditions. Here, we identified a mechanism by which HDAC3 regulates DNA damage repair and mediates resistance to chemotherapy drugs. In addition to inducing DNA damage, chemotherapy drugs trigger upregulation of HDAC3 expression in leukemia cells. Using genetic and pharmacological approaches, we show that HDAC3 contributes to chemotherapy resistance by regulating the activation of AKT, a well-documented factor in drug resistance development. HDAC3 binds to AKT and deacetylates it at the site Lys20, thereby promoting the phosphorylation of AKT. Chemotherapy drug exposure enhances the interaction between HDAC3 and AKT, resulting in decrease in AKT acetylation and increase in AKT phosphorylation. Whereas HDAC3 depletion or inhibition abrogates these responses and meanwhile sensitizes leukemia cells to chemotoxicity-induced apoptosis. Importantly, in vivo HDAC3 suppression reduces leukemia progression and sensitizes MLL-AF9+ leukemia to chemotherapy. Our findings suggest that combination therapy with HDAC3 inhibitor and genotoxic agents may constitute a successful strategy for overcoming chemotherapy resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Dohner H, Weisdorf DJ, Bloomfield CD . Acute myeloid leukemia. N Engl J Med 2015; 373: 1136–1152.
Ferrara F, Schiffer CA . Acute myeloid leukaemia in adults. Lancet 2013; 381: 484–495.
Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M . Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood 2003; 102: 972–980.
Grandage VL, Gale RE, Linch DC, Khwaja A . PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia 2005; 19: 586–594.
Min YH, Eom JI, Cheong JW, Maeng HO, Kim JY, Jeung HK et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia 2003; 17: 995–997.
West KA, Castillo SS, Dennis PA . Activation of the PI3K/Akt pathway and chemotherapeutic resistance. Drug Resist Updat 2002; 5: 234–248.
O'Gorman DM, McKenna SL, McGahon AJ, Knox KA, Cotter TG . Sensitisation of HL60 human leukaemic cells to cytotoxic drug-induced apoptosis by inhibition of PI3-kinase survival signals. Leukemia 2000; 14: 602–611.
Brandwein JM, Hedley DW, Chow S, Schimmer AD, Yee KW, Schuh AC et al. A phase I/II study of imatinib plus reinduction therapy for c-kit-positive relapsed/refractory acute myeloid leukemia: inhibition of Akt activation correlates with complete response. Leukemia 2011; 25: 945–952.
Bozulic L, Surucu B, Hynx D, Hemmings BA . PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell 2008; 30: 203–213.
Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FM . Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 2015; 15: 166–180.
Roos WP, Thomas AD, Kaina B . DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 2016; 16: 20–33.
West AC, Johnstone RW . New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014; 124: 30–39.
Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell 2006; 9: 249–260.
Lahue RS, Frizzell A . Histone deacetylase complexes as caretakers of genome stability. Epigenetics 2012; 7: 806–810.
Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C . Histone deacetylase inhibitors and genomic instability. Cancer Lett 2009; 274: 169–176.
Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010; 18: 436–447.
Lane AA, Chabner BA . Histone deacetylase inhibitors in cancer therapy. J Clin Oncol 2009; 27: 5459–5468.
Yang CZ, Luan FJ, Xiong DS, Liu BR, Xu YF, Gu KS . Multidrug resistance in leukemic cell line K562/A02 induced by doxorubicin. Zhongguo Yao Li Xue Bao 1995; 16: 333–337.
Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev 2009; 23: 877–889.
Long J, Chang L, Shen Y, Gao WH, Wu YN, Dou HB et al. Valproic acid ameliorates graft-versus-host disease by downregulating Th1 and Th17 cells. J Immunol 2015; 195: 1849–1857.
Toulany M, Kehlbach R, Florczak U, Sak A, Wang S, Chen J et al. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther 2008; 7: 1772–1781.
Krivtsov AV, Armstrong SA . MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007; 7: 823–833.
Milne TA . Mouse models of MLL leukemia: recapitulating the human disease. Blood 2017; 129: 2217–2223.
Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J et al. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol 2015; 33: 415–423.
Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008; 30: 61–72.
Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal 2011; 4: ra46.
Watson PJ, Fairall L, Santos GM, Schwabe JW . Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 2012; 481: 335–340.
Arrar M, de Oliveira CA, McCammon JA . Inactivating mutation in histone deacetylase 3 stabilizes its active conformation. Protein Sci 2013; 22: 1306–1312.
Adams H, Fritzsche FR, Dirnhofer S, Kristiansen G, Tzankov A . Class I histone deacetylases 1, 2 and 3 are highly expressed in classical Hodgkin's lymphoma. Expert Opin Ther Targets 2010; 14: 577–584.
Muller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer—overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer 2013; 13: 215.
Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V et al. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol 2010; 150: 665–673.
Wang JC, Kafeel MI, Avezbakiyev B, Chen C, Sun Y, Rathnasabapathy C et al. Histone deacetylase in chronic lymphocytic leukemia. Oncology 2011; 81: 325–329.
Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A . Dysregulated class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics 2014; 9: 1511–1520.
Quintas-Cardama A, Santos FP, Garcia-Manero G . Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia 2011; 25: 226–235.
Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA non-homologous end-joining. Nat Struct Mol Biol 2010; 17: 1144–1151.
Groselj B, Sharma NL, Hamdy FC, Kerr M, Kiltie AE . Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. Br J Cancer 2013; 108: 748–754.
Xu N, Lao Y, Zhang Y, Gillespie DA . Akt: a double-edged sword in cell proliferation and genome stability. J Oncol 2012; 2012: 951724.
Toulany M, Lee KJ, Fattah KR, Lin YF, Fehrenbacher B, Schaller M et al. Akt promotes post-irradiation survival of human tumor cells through initiation, progression, and termination of DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Res 2012; 10: 945–957.
Deng R, Tang J, Ma JG, Chen SP, Xia LP, Zhou WJ et al. PKB/Akt promotes DSB repair in cancer cells through upregulating Mre11 expression following ionizing radiation. Oncogene 2011; 30: 944–955.
Brown KK, Montaser-Kouhsari L, Beck AH, Toker A . MERIT40 is an Akt substrate that promotes resolution of DNA damage induced by chemotherapy. Cell Rep 2015; 11: 1358–1366.
Gupta M, Ansell SM, Novak AJ, Kumar S, Kaufmann SH, Witzig TE . Inhibition of histone deacetylase overcomes rapamycin-mediated resistance in diffuse large B-cell lymphoma by inhibiting Akt signaling through mTORC2. Blood 2009; 114: 2926–2935.
Bradley EW, Carpio LR, Westendorf JJ . Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem 2013; 288: 9572–9582.
Yang XJ, Seto E . Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 2008; 31: 449–461.
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325: 834–840.
Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev 2009; 23: 223–235.
Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE . Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia 2012; 26: 1356–1364.
Minami J, Suzuki R, Mazitschek R, Gorgun G, Ghosh B, Cirstea D et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia 2014; 28: 680–689.
Rebecchi MJ, Scarlata S . Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct 1998; 27: 503–528.
Matthews GM, Mehdipour P, Cluse LA, Falkenberg KJ, Wang E, Roth M et al. Functional-genetic dissection of HDAC dependencies in mouse lymphoid and myeloid malignancies. Blood 2015; 126: 2392–2403.
Acknowledgements
We thank all fellows from Research Center for Experimental Medicine for their technical assistance. In addition, we thank all researchers from Shanghai Institute of Hematology for kind advice and the supply of research reagents. This Study is supported by the Ministry of Science and Technology of China (S2016G9074) and National Natural Science Foundation of China (81270615, 81270621, 81123005).
Author contributions
JL, WYF and LC performed the experiments, analyzed the data and helped to write the manuscript; WHG performed experiments and helped with animal experiments; YS and MYJ helped with animal experiments; YXZ helped to collect primary samples; YW, HBD and WJZ provided advice and revised the paper; JZ provided material and advice, and revised the paper; ABL, JML and JH designed the overall concept, analyzed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Long, J., Fang, W., Chang, L. et al. Targeting HDAC3, a new partner protein of AKT in the reversal of chemoresistance in acute myeloid leukemia via DNA damage response. Leukemia 31, 2761–2770 (2017). https://doi.org/10.1038/leu.2017.130
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.130
This article is cited by
-
Post-translational modification of CDK1–STAT3 signaling by fisetin suppresses pancreatic cancer stem cell properties
Cell & Bioscience (2023)
-
Targeting HDAC3 to overcome the resistance to ATRA or arsenic in acute promyelocytic leukemia through ubiquitination and degradation of PML-RARα
Cell Death & Differentiation (2023)
-
Disruption of mitochondrial oxidative phosphorylation by chidamide eradicates leukemic cells in AML
Clinical and Translational Oncology (2023)
-
HDAC11 mediates the ubiquitin-dependent degradation of p53 and inhibits the anti-leukemia effect of PD0166285
Medical Oncology (2023)
-
The NCOR-HDAC3 co-repressive complex modulates the leukemogenic potential of the transcription factor ERG
Nature Communications (2023)