Abstract
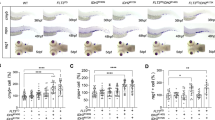
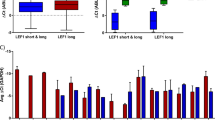
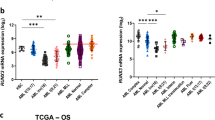
cAMP response element binding protein (CREB) is frequently overexpressed in acute myeloid leukemia (AML) and acts as a proto-oncogene; however, it is still debated whether such overactivation alone is able to induce leukemia as its pathogenetic downstream signaling is still unclear. We generated a zebrafish model overexpressing CREB in the myeloid lineage, which showed an aberrant regulation of primitive hematopoiesis, and in 79% of adult CREB-zebrafish a block of myeloid differentiation, triggering to a monocytic leukemia akin the human counterpart. Gene expression analysis of CREB-zebrafish revealed a signature of 20 differentially expressed human homologous CREB targets in common with pediatric AML. Among them, we demonstrated that CREB overexpression increased CCAAT-enhancer-binding protein-δ (C/EBPδ) levels to cause myeloid differentiation arrest, and the silencing of CREB-C/EBPδ axis restored myeloid terminal differentiation. Then, C/EBPδ overexpression was found to identify a subset of pediatric AML affected by a block of myeloid differentiation at monocytic stage who presented a significant higher relapse risk and the enrichment of aggressive signatures. Finally, this study unveils the aberrant activation of CREB-C/EBPδ axis concurring to AML onset by disrupting the myeloid cell differentiation process. We provide a novel in vivo model to perform high-throughput drug screening for AML cure improvement.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 2005; 7: 351–362.
Sandoval S, Pigazzi M, Sakamoto KM . CREB: a key regulator of normal and neoplastic hematopoiesis. Adv Hematol 2009; 2009: 634292.
Pigazzi M, Manara E, Baron E, Basso G . ICER expression inhibits leukemia phenotype and controls tumor progression. Leukemia 2008; 22: 2217–2225.
Sandoval S, Kraus C, Cho EC, Cho M, Bies J, Manara E et al. Sox4 cooperates with CREB in myeloid transformation. Blood 2012; 120: 155–165.
Pigazzi M, Ricotti E, Germano G, Faggian D, Aricò M, Basso G . cAMP response element binding protein (CREB) overexpression CREB has been described as critical for leukemia progression. Haematologica 2007; 92: 1435–1437.
Crans-Vargas HN, Landaw EM, Bhatia S, Sandusky G, Moore TB, Sakamoto KM . Expression of cyclic adenosine monophosphate response-element binding protein in acute leukemia. Blood 2002; 99: 2617–2619.
Pigazzi M, Manara E, Beghin A, Baron E, Tregnago C, Basso G . ICER evokes Dusp1-p38 pathway enhancing chemotherapy sensitivity in myeloid leukemia. Clin Cancer Res 2011; 17: 742–752.
Pigazzi M, Manara E, Baron E, Basso G . MiR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res 2009; 69: 2471–2478.
Pigazzi M, Manara E, Bresolin S, Tregnago C, Beghin A, Baron E et al. MicroRNA-34b promoter hypermethylation induces CREB overexpression and contributes to myeloid transformation. Haematologica 2013; 98: 602–610.
Pui C-H, Carroll WL, Meshinchi S, Arceci RJ . Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol 2011; 29: 551–565.
Pession A, Masetti R, Rizzari C, Putti MC, Casale F, Fagioli F et al. Results of the AIEOP AML 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood 2013; 122: 170–178.
Gibson BES, Webb DKH, Howman AJ, De Graaf SSN, Harrison CJ, Wheatley K . Results of a randomized trial in children with Acute Myeloid Leukaemia: medical research council AML12 trial. Br J Haematol 2011; 155: 366–376.
Abrahamsson J, Forestier E, Heldrup J, Jahnukainen K, Jónsson OG, Lausen B et al. Response-guided induction therapy in pediatric acute myeloid leukemia with excellent remission rate. J Clin Oncol 2011; 29: 310–315.
Ribeiro RC . Advances in treatment of de-novo pediatric acute myeloid leukemia. Curr Opin Oncol 2014; 26: 656–662.
Pigazzi M, Masetti R, Martinolli F, Manara E, Beghin A, Rondelli R et al. Presence of high-ERG expression is an independent unfavorable prognostic marker in MLL-rearranged childhood myeloid leukemia. Blood 2012; 119: 1086–1087; author reply 1087–1088.
Manara E, Bisio V, Masetti R, Beqiri V, Rondelli R, Menna G et al. Core-binding factor acute myeloid leukemia in pediatric patients enrolled in the AIEOP AML 2002/01 trial: screening and prognostic impact of c-KIT mutations. Leukemia 2014; 28: 1132–1134.
Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood 2009; 114: 2489–2496.
Vassal G, Zwaan CM, Ashley D, Le Deley MC, Hargrave D, Blanc P et al. New drugs for children and adolescents with cancer: the need for novel development pathways. Lancet Oncol 2013; 14: e117–e124.
Bautista F, Di Giannatale A, Dias-Gastellier N, Fahd M, Valteau-Couanet D, Couanet D et al. Patients in pediatric phase I and early phase II clinical oncology trials at Gustave Roussy: a 13-year center experience. J Pediatr Hematol Oncol 2015; 37: e102–e110.
Santoriello C, Zon LI . Hooked! Modeling human disease in zebrafish. J Clin Invest 2012; 122: 2337–2343.
Jing L, Zon LI . Zebrafish as a model for normal and malignant hematopoiesis. Dis Model Mech 2011; 4: 433–438.
Liu S, Leach SD . Zebrafish models for cancer. Annu Rev Pathol 2011; 6: 71–93.
Davidson AJ, Zon LI . The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 2004; 23: 7233–7246.
Langenau DM, Traver D, Ferrando Aa, Kutok JL, Aster JC, Kanki JP et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003; 299: 887–890.
Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med 2011; 208: 1595–1603.
Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD . NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 2007; 21: 462–471.
Sabaawy HE, Azuma M, Embree LJ, Tsai H-J, Starost MF, Hickstein DD . TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA 2006; 103: 15166–15171.
Zhuravleva J, Paggetti J, Martin L, Hammann A, Solary E, Bastie JN et al. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br J Haematol 2008; 143: 378–382.
Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT . Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol 2009; 5: 236–243.
Gutierrez A, Pan L, Groen RWJ, Baleydier F, Kentsis A, Marineau J et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest 2014; 124: 644–655.
Dworkin S, Heath JK, deJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J et al. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev Biol 2007; 307: 127–141.
Wilcoxon F . Individual comparisons of grouped data by ranking methods. J Econ Entomol 1946; 39: 269.
Haferlach T, Kohlmann A, Wieczorek L, Basso G, Te Kronnie G, Béné MC et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the international microarray innovations in leukemia study group. J Clin Oncol 2010; 28: 2529–2537.
Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP . GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007; 23: 3251–3253.
Hoshida Y . Nearest Template Prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One 2010; 5: e15543.
Hsu K, Traver D, Kutok JL, Hagen A, Liu T-X, Paw BH et al. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood 2004; 104: 1291–1297.
Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D . Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 2007; 134: 4147–4156.
Zhang X, Odom DT, Koo S-H, Conkright MD, Canettieri G, Best J et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 2005; 102: 4459–4464.
Alberich-Jordà M, Wouters B, Balastik M, Shapiro-Koss C, Zhang H, DiRuscio A et al. C/EBPγ deregulation results in differentiation arrest in acute myeloid leukemia. J Clin Invest 2012; 122: 4490–4504.
Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet 2001; 27: 263–270.
Hirai H, Yokota A, Tamura A, Sato A, Maekawa T . Non-steady-state hematopoiesis regulated by the C/EBPβ transcription factor. Cancer Sci 2015; 106: 797–802.
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol 1976; 33: 451–458.
Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci USA 2009; 106: 12944–12949.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39: 232–236.
Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008; 454: 49–55.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP . Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27: 1739–1740.
Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR . Identification of direct genomic targets downstream of the nuclear factor-κB transcription factor mediating tumor necrosis factor signaling. J Biol Chem 2005; 280: 17435–17448.
Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol 2007; 27: 4374–4387.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357.
Gary Gilliland D, Griffin JD . The roles of FLT3 in hematopoiesis and leukemia. Blood 2002; 100: 1532–1542.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837.
Acknowledgements
We thank Dr Patrizia Porazzi for zebrafish manipulation, Dr Benedetta Accordi and Dr Valentina Serafin for RPPA analysis, Dr Chiara Frasson for cell sorting and Dr Luca Persano for tissue sectioning. This work was supported by grants from Cariparo, IRP-Istituto di Ricerca Pediatrica Città della Speranza Padova and Università degli Studi di Padova.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Tregnago, C., Manara, E., Zampini, M. et al. CREB engages C/EBPδ to initiate leukemogenesis. Leukemia 30, 1887–1896 (2016). https://doi.org/10.1038/leu.2016.98
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.98
This article is cited by
-
Zebrafish: a convenient tool for myelopoiesis research
Cell Regeneration (2023)
-
Detecting the stable point of therapeutic effect of chronic myeloid leukemia based on dynamic network biomarkers
BMC Bioinformatics (2019)
-
The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia
Molecular Cancer (2017)
-
Overexpression of SOX4 correlates with poor prognosis of acute myeloid leukemia and is leukemogenic in zebrafish
Blood Cancer Journal (2017)