Abstract
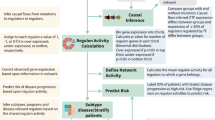
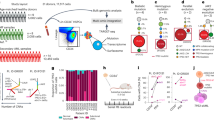
TP53 mutations are associated with the lowest survival rates in acute myeloid leukemia (AML). In addition to mutations, loss of p53 function can arise via aberrant expression of proteins that regulate p53 stability and function. We examined a large AML cohort using proteomics, mutational profiling and network analyses, and showed that (1) p53 stabilization is universal in mutant TP53 samples, it is frequent in samples with wild-type TP53, and in both cases portends an equally dismal prognosis; (2) the p53 negative regulator Mdm2 is frequently overexpressed in samples retaining wild-type TP53 alleles, coupled with absence of p21 expression and dismal prognosis similar to that of cases with p53 stabilization; (3) AML samples display unique patterns of p53 pathway protein expression, which segregate prognostic groups with distinct cure rates; (4) such patterns of protein activation unveil potential AML vulnerabilities that can be therapeutically exploited.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Vogelstein B, Kinzler KW . Cancer genes and the pathways they control. Nat Med 2004; 10: 789–799.
Oren M . Decision making by p53: life, death and cancer. Cell Death Differ 2003; 10: 431–442.
Hollstein M, Sidransky D, Vogelstein B, Harris CC . p53 mutations in human cancers. Science 1991; 253: 49–53.
Vogelstein B, Lane D, Levine AJ . Surfing the p53 network. Nature 2000; 408: 307–310.
Stolzel F, Pfirrmann M, Aulitzky WE, Kaufmann M, Bodenstein H, Bornhauser M et al. Risk stratification using a new prognostic score for patients with secondary acute myeloid leukemia: results of the prospective AML96 trial. Leukemia 2011; 25: 420–428.
Seifert H, Mohr B, Thiede C, Oelschlagel U, Schakel U, Illmer T et al. The prognostic impact of 17p (p53) deletion in 2272 adults with acute myeloid leukemia. Leukemia 2009; 23: 656–663.
Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Nahi H, Lehmann S, Bengtzen S, Jansson M, Mollgard L, Paul C et al. Chromosomal aberrations in 17p predict in vitro drug resistance and short overall survival in acute myeloid leukemia. Leuk Lymphoma 2008; 49: 508–516.
Kubbutat MH, Jones SN, Vousden KH . Regulation of p53 stability by Mdm2. Nature 1997; 387: 299–303.
Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 1998; 92: 713–723.
Wasylishen AR, Lozano G . Attenuating the p53 pathway in human cancers: many means to the same end. Cold Spring Harb Perspect Med 2016; 6: a026211.
Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B . Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992; 358: 80–83.
Momand J, Jung D, Wilczynski S, Niland J . The MDM2 gene amplification database. Nucleic Acids Res 1998; 26: 3453–3459.
Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G . An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 2001; 20: 4041–4049.
Kornblau SM, Womble M, Qiu YH, Jackson CE, Chen W, Konopleva M et al. Simultaneous activation of multiple signal transduction pathways confers poor prognosis in acute myelogenous leukemia. Blood 2006; 108: 2358–2365.
Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M et al. Functional proteomic profiling of AML predicts response and survival. Blood 2009; 113: 154–164.
Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther 2006; 5: 2512–2521.
Hunyady B, Krempels K, Harta G, Mezey E . Immunohistochemical signal amplification by catalyzed reporter deposition and its application in double immunostaining. J Histochem Cytochem 1996; 44: 1353–1362.
Neeley ES, Kornblau SM, Coombes KR, Baggerly KA . Variable slope normalization of reverse phase protein arrays. Bioinformatics 2009; 25: 1384–1389.
Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB . Non-parametric quantification of protein lysate arrays. Bioinformatics 2007; 23: 1986–1994.
Neeley ES, Baggerly KA, Kornblau SM . Surface adjustment of reverse phase protein arrays using positive control spots. Cancer Inform 2012; 11: 77–86.
Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z et al. A pan-cancer proteomic perspective on the Cancer Genome Atlas. Nat Commun 2014; 5: 3887.
Hu CW, Kornblau SM, Slater JH, Qutub AA . Progeny clustering: a method to identify biological phenotypes. Sci Rep 2015; 5: 12894.
Hu CYW, Kornblau SM, Slater JH, Qutub AA . Progeny clustering: a method to identify biological phenotypes. Sci Rep-UK 2015; 5: 12894.
Tibshirani R . The lasso method for variable selection in the Cox model. Stat Med 1997; 16: 385–395.
Friedman J, Hastie T, Tibshirani R . Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008; 9: 432–441.
Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci USA 2009; 106: 11612–11616.
Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998; 281: 1674–1677.
Shieh SY, Ikeda M, Taya Y, Prives C . DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91: 325–334.
Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 2002; 21: 2613–2622.
Yang HY, Wen YY, Lin YI, Pham L, Su CH, Yang H et al. Roles for negative cell regulator 14-3-3sigma in control of MDM2 activities. Oncogene 2007; 26: 7355–7362.
Sauerbrey A, Stammler G, Zintl F, Volm M . Expression of the retinoblastoma tumor suppressor gene (RB-1) in acute leukemia. Leuk Lymphoma 1998; 28: 275–283.
Melo MB, Costa FF, Saad ST, Lorand-Metze I, Bordin S, Ahmad NN . Molecular analysis of the retinoblastoma (RB1) gene in acute myeloid leukemia patients. Leuk Res 1998; 22: 787–792.
Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 2010; 24: 1389–1402.
Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL et al. p14ARF links the tumour suppressors RB and p53. Nature 1998; 395: 124–125.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998; 12: 2424–2433.
Winter PS, Sarosiek KA, Lin KH, Meggendorfer M, Schnittger S, Letai A et al. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci Signal 2014; 7: ra122.
Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal 2014; 7: ra121.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Quintás-Cardama, A., Hu, C., Qutub, A. et al. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia 31, 1296–1305 (2017). https://doi.org/10.1038/leu.2016.350
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.350
This article is cited by
-
p53 biology and reactivation for improved therapy in MDS and AML
Biomarker Research (2024)
-
Translating p53-based therapies for cancer into the clinic
Nature Reviews Cancer (2024)
-
Logic-based modeling and drug repurposing for the prediction of novel therapeutic targets and combination regimens against E2F1-driven melanoma progression
BMC Chemistry (2023)
-
miR-221/222 induce instability of p53 By downregulating deubiquitinase YOD1 in acute myeloid leukemia
Cell Death Discovery (2023)
-
Extracellular vesicles transfer chromatin-like structures that induce non-mutational dysfunction of p53 in bone marrow stem cells
Cell Discovery (2023)