Abstract
Heterozygous somatic mutations affecting the spliceosome gene SF3B1 drive age-related clonal hematopoiesis, myelodysplastic syndromes (MDS) and other neoplasms. To study their role in such disorders, we generated knock-in mice with hematopoietic-specific expression of Sf3b1-K700E, the commonest type of SF3B1 mutation in MDS. Sf3b1K700E/+ animals had impaired erythropoiesis and progressive anemia without ringed sideroblasts, as well as reduced hematopoietic stem cell numbers and host-repopulating fitness. To understand the molecular basis of these observations, we analyzed global RNA splicing in Sf3b1K700E/+ hematopoietic cells. Aberrant splicing was associated with the usage of cryptic 3′ splice and branchpoint sites, as described for human SF3B1 mutants. However, we found a little overlap between aberrantly spliced mRNAs in mouse versus human, suggesting that anemia may be a consequence of globally disrupted splicing. Furthermore, the murine orthologues of genes associated with ring sideroblasts in human MDS, including Abcb7 and Tmem14c, were not aberrantly spliced in Sf3b1K700E/+ mice. Our findings demonstrate that, despite significant differences in affected transcripts, there is overlap in the phenotypes associated with SF3B1-K700E between human and mouse. Future studies should focus on understanding the basis of these similarities and differences as a means of deciphering the consequences of spliceosome gene mutations in MDS.
Similar content being viewed by others
Introduction
The myelodysplastic syndromes (MDS) are clonal blood disorders characterized by dysplastic hematopoiesis and blood cytopenias.1 Somatic mutations affecting the genes SF3B1, SRSF2, U2AF1, ZRSR2 and LUC7L2, which code for components of the spliceosome, are found in more than half of MDS patients.2, 3 SF3B1 mutations are the commonest among these and are associated with the presence of bone marrow ringed sideroblasts2 and a favorable prognosis.4, 5 The mutations are missense, heterozygous and cluster strongly within HEAT domains 4–8 of the protein, suggesting that they may be neomorphic gain-of-function variants.2 Studies analyzing the subclonal composition of MDS indicate that SF3B1 mutations represent early/initiating lesions in MDS evolution,5, 6, 7 and this was corroborated by their recent identification in hematopoietic cell clones found in at least 2% of otherwise healthy individuals aged 70 years or older.8 Furthermore, in a significant proportion of SF3B1-mutant MDS this is the only identifiable driver mutation,7, 9 suggesting that in the right context these mutations can impart a substantial fitness advantage on hematopoietic stem cells. However, the molecular effects of SF3B1 mutations and the mechanisms through which they drive clonal expansion and dyserythropoiesis remain obscure.
RNA splicing takes place at splice sites (ss) located at the 5′ and 3′ ends of introns, after the recognition of consensus nucleotide sequences by the spliceosome machinery. The U1 small nuclear ribonucleoprotein particle (snRNP) recognizes the 5′ ss, and at the 3′ ss the polypyrimidine (Py) tract and an invariant AG dinucleotide located at the intron-exon junction are recognized by the U2AF1–U2AF2 complex, while the proximal branch point sequence (BPS) is identified by the U2 snRNP, of which SF3B1 is a component.10 MDS cases associated with SF3B1 mutations show a distinct gene expression profile, including dysregulation of genes required for heme biosynthesis, such as ALAS2 and ABCB7.11, 12 Furthermore, it was recently demonstrated in primary human cancers and cell lines that SF3B1 mutations are associated with aberrant splicing through recognition of alternative 3′ ss located around 10–25 nt upstream of the canonical 3′ ss.13 However, the consequences of these aberrations and their impact on hematopoiesis are not well understood.
Here, to investigate the molecular and phenotypic consequences of SF3B1 mutations on hematopoiesis, we generate and study an accurate conditional knock-in mouse model of SF3B1-K700E, the commonest mutation in MDS. Our findings show that while the sequence features of the aberrant splicing are extremely similar between mouse and human,13 the genes affected differ significantly. Despite these marked differences, Sf3b1K700E/+ mice did go on to develop progressive anemia, a central feature of SF3B1-mutant MDS. However, other characteristic features of SF3B1-mutant MDS such as ring sideroblasts and a stem cell growth advantage were not observed. Our results propose that the anemia associated with mutant SF3B1 may be a consequence of globally disrupted splicing rather than effects on particular genes while other features of SF3B1-mutant MDS may be gene-specific.
Materials and methods
Generation of the Sf3b1-K700E targeting construct
The targeting construct was generated using gateway and recombineering recombination technologies as described previously.14 Briefly, a pool of two BACs containing the region of interest of the Sf3b1 locus, RP24-64H9 and RP24-439B17, were used to generate the gateway-adapted intermediate plasmid by inserting the ‘U’ and ‘G’ cassettes.14 The latter were generated by PCR and the ‘U’ cassette inserted into the BAC first so that the ‘G’ cassette then retrieved the relevant portion of the Sf3b1 locus into a gateway-adapted intermediate plasmid.15 The endogenous exon 15 was then replaced with a synthetic one bearing the A>G mutation encoding for the K700E variant. The synthetic DNA was made by GeneArt (Life Technologies, Regensburg, Germany), and was subcloned into the intermediate vector exploiting two KpnI sites in introns 14 and 16 (Supplementary Figure 1). The splice trap cassette encoding an Engrail-2 splice acceptor site, codon-optimized exons 12–15, intron 15, codon-optimized exons 15–19, intron 19 and native exons 20–25 (including the 3′ UTR) was synthesized by GeneArt (Life Technologies) and subcloned into the pL1L2_BactP vector previously modified to remove the EnSA-IRES-lacZ-pA-LoxP and subsequently the 3′ LoxP site by digestion. The final targeting vector was built using the two vectors above and the pL3L4-DTA vector14 in a multi-vector gateway reaction, and was linearized prior to electroporation using the AsisI restriction site (Supplementary Figure 2). Sequences of primers used to generate and validate the targeting construct are given in Supplementary Table 1.
ES cell culture and validation of clones
JM8 ES cells were used for electroporation of the final targeting vector. Cells were grown for 9 days after electroporation in a feeder-coated plate with DMEM medium (Invitrogen, Hempstead, UK) supplemented with 10% FBS (Biosera, UK), 1% BME (Sigma-Aldrich, Haverhill, UK), 1000 U/ml leukemia inhibitory factor (ESGRO, Millipore, UK) and G418 (Sigma-Aldrich) 180 μg/ml. Clones were picked and replated in a 96-well plate with feeders, split in replica plates after 5 days, and lysis for DNA extraction was performed after seven additional days. Clones were screened by 5′ and 3′ long-range PCR (Supplementary Figure 3). Three clones with successful 5′ and 3′ amplification, were and all showed that the splice-trap cassette was indeed functional (Supplementary Figure 4a and b). ES cell clones post exposure to FLP and Cre recombinases were validated for excision of the positive selection marker and cDNA splice-trap cassette, respectively (Supplementary Figure 4c). To confirm that the K700E mutation was not expressed in basal conditions but only after Cre expression, cDNA from clone D9 pre- and post- excision of the splice trap cassette was sequenced (Supplementary Figure 4d).
Hematological measurements and mouse survival
Blood counts were performed using a VetABC analyzer (Horiba ABX, Montpellier, France). Mice were monitored daily and culled if they showed signs of illness or suffering. All animal studies were performed according to the Animals Scientific Procedures Act 1986 (ASPA), as recently revised to transpose European Directive 2010/63/EU on the protection of animals used for scientific purposes. Mouse cohort numbers were estimated to detect (P <0.05) a difference in survival of 2 months or greater with a power of 0.8, assuming a standard deviation of 2.2 months (http://www.biomath.info/power/ttest.htm).
Competitive hematopoietic progenitor transplantation
C57BL/6 mice (n=4 per group) were irradiated with a lethal dose of 2 × 500 rad. They were then injected into the tail vein with a combination of 250 000 lineage negative bone marrow cells (lin−) from C57BL/6.SJL (Sf3b1+/+, CD45.1) mice and 250 000 lin− cells from Sf3b1K700E/+ (CD45.2) mice plus 250 000 whole bone marrow (BM) rescue cells from C57BL/6/6.SJL (CD45.1/CD45.2) mice. Control animals were injected with 1 × 106 whole BM cells from C57BL/6/6.SJL (CD45.1/CD45.2). Blood counts and FACS analysis were performed after 1, 2 and 4 months. Samples were stained with CD45.1-APC (Becton Dickinson, Oxford, UK) and CD45.2-FITC (Becton Dickinson) antibodies run on Becton Dickinson LSFR Fortessa and analyzed with Flowjo 7.6.5 (FlowJo, LLC, San Diego, CA, USA).
Immunophenotyping
For mouse phenotyping peripheral blood and bone marrow from femurs were collected from animals of different genotypes and processed as previously described.16 For flow analysis, samples were stained with Gr1-PE (Miltenyi Biotec, Woking, UK), CD11b (Mac1)-FITC (Becton Dickinson), CD71-PE (Becton Dickinson), Ter119-Fitc (Becton Dickinson). For analysis of progenitor populations BM cells were lysed with Red Blood Cell buffer (NH4, Cl) and then enriched with the EasySep Hematopoietic Progenitor Cell Isolation Kit (StemCell Technologies, Cambridge, UK) followed by staining with following antibodies Sca1-PB, c-KIT-Acy7, CD34-FITC, Flk2-PeCy5, Cd16/32-PE, CD150-PeCy7, CD48-APC.
RNA isolation and analysis
Whole BM cells were harvested from femurs of Sf3b1+/+ and Sf3b1K700E/+ animals. lin− cells were isolated with MACS separation system (Miltenyi Biotec) following standard protocol. RNA from whole BM was isolated using an RNA isolation Kit (Qiagen, Manchester, UK) and RNA from lin− cells with PicoPure RNA Isolation System (Thermo Fisher, Hempstead, UK). Sequencing was performed on the Hiseq2000 V4 sequencing platform (llumina, San Diego, CA, USA). Sequence data have been deposited in ArrayExpress with the accession number E-ERAD-379. All RNA-seq data were analyzed as described previously.13 RNA-seq datasets have been deposited at the NCBI GEO and are available under accession number GEO:GSE72790. STAR alignment software was used to align raw sequence fragments to the hg19 human reference genome.17 Junction percent spliced-in (PSI) was calculated for any two or more splice junctions that share a junction boundary and have alternative end points by dividing the raw count of each junction species by the total sum of junction counts that also share that boundary. Intron retention was factored into PSI calculations by counting reads that aligned completely to exon–intron boundaries, defined as 6 basepair (bp) windows, 3 bp in the exon and 3 bp in the intron. We used a moderated t-test from the stastical package limma18 to determine the significance of PSI measurements. We used the Gencode v19 transcriptome annotation to evaluate whether junction boundaries were equivalent to known exon boundaries (that is, junction novelty). Gene counts were obtained using the Sailfish package,19 and differential gene expression calculated using edgeR.20 All P-values were corrected using the Benjamini–Hochberg procedure and q-values <0.05 were considered significant.
Nested PCR for lariat sequencing
Gene-specific reverse transcription of the endogenous lariat region was done using Superscript IV (Invitrogen) and primer C (Supplementary Table 2). The resulting cDNA was used for nested PCR reactions using Platinum Taq DNA polymerase (Invitrogen) and two sets of primers, outer primers (C and D) followed by inner primers (A and B) (Supplementary File 3). The PCR product from the second reaction was run on 2.0% agarose gel and gel-extracted for Sanger sequencing. The sequencing data were analyzed using Mutation Surveyor software.
Statistical analyses
Blood counts and cellular compartment sizes were compared using unpaired t-test. Mouse survival was compared using the Kaplan Meier estimator. Adenine counts near canonical vs aberrant splice junctions and expression levels of transcripts that were NMD-predicted vs non-NMD predicted were compared using the Kruskal H test (Figure 3). Comparison of RNA-seq transcript levels was performed using edgeR20 (Figure 4).
Aberrant 3′ splicing is the dominant abnormality associated with Sf3b1-K700E in bone marrow cells. (a) Types and relative frequencies of aberrant splicing events in Sf3b1K700E/+ vs WT BM and lin− cells. Most are alternative 3′ splicing events, of which many are shared between BM and lin− cells (Venn diagram). (b) Distribution of distances of aberrant 3′ss (AG) from the downstream canonical 3′ss in Sf3b1K700E/+ BM and lin− RNA. This distance is significantly shorter than that of known cryptic ss annotated in Ensembl (all mouse junctions for the first AAG, TAG or CAG within 100 nt upstream from the canonical 3' splice site are considered). (c) Motif frequency plots for Sf3b1K700E/+ BM and lin− samples contrasting aberrant and canonical 3′ ss sequences, showing an enrichment of adenines in the former. (d) Significant enrichment in adenines in positions -8 to -18 upstream of aberrant 3′ ss (AG) compared to the canonical junctions from Sf3b1K700E/+ BM and lin− cells. (e) Lariat sequencing of the aberrant splice site of the Ugdh gene in Sf3b1K700E/+ mice, highlighting the location of the aberrant branch point (upper panel; the top trace is the raw data from Sanger sequencing and bottom is deconvoluted data from Mutation Surveyor). The gene is not misspliced at this locus in human SF3B1-mutant cancers as it does not retain the AG dinucleotide required for aberrant splicing (lower panel). (f) Association between aberrant splicing and gene expression. Transcripts that are NMD-predicted are significantly downregulated compared to transcripts that are non-NMD predicted (p = 3.92 × 10−13, Kruskal H test).
Mouse and human SF3B1-K700E share aberrant splicing properties, but affect different mRNA transcripts. (a) Comparison of consensus sequences upstream of aberrant and canonical 3′ ss shows a striking preservation of these motifs between mouse Sf3b1K700E/+ lin− cells and several human SF3B1-mutant cancers. (b) Distribution of distances of aberrant 3′ss (AG) from the downstream canonical AG is also strikingly similar between mouse Sf3b1K700E/+ lin− cells and human cancer samples. (c) By contrast, a comparison of genes found to be aberrantly spliced in human vs mouse samples shows minimal overlap. (d) Poor sequence preservation in the mouse of the aberrant splice sites affecting human genes TMEM14C and ABCB7 that are thought to trigger the formation of ring sideroblasts in MDS. (e) Volcano plots comparing mRNA expression between Sf3b1K700E/+ vs Sf3b1+/+ BM and lin− cells. The mRNA expression of genes Slc25a37, Tmem14c, Alas2 and Abcb7, thought to be involved in the formation of ring sideroblasts, is not significantly altered by the K700E mutation in mice. The number of aberrant events used to generate images in a and b is indicated in b (brackets). BRCA, breast cancer; CLL, chronic lymphocytic leukemia; MDS, myelodysplastic syndromes; SKCM, skin cutaneous melanoma.
Results
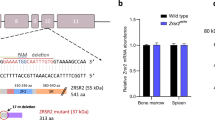
Conditional Sf3b1flox-K700E/+ mice were generated by targeted modification of the Sf3b1 locus in JM8 mouse embryonic stem cells (ESCs)14, 21 (Figure 1a and Supplementary Figures 1 and 2). To ensure that normal mRNA splicing was retained, the targeted allele was designed to express normal SF3B1 protein via a part-codon optimized Sf3b1 exon 12-25 cDNA (Figure 1a and Supplementary Figure 3). After germline transmission, a downstream neomycin cassette was removed in vivo through mating with Rosa26-FLPe mice,22 to generate the conditional Sf3b1flox−K700E allele (Supplementary Figure 3). The Sf3b1K700E mutant allele could then be expressed after Cre-loxP recombination (Figure 1a and Supplementary Figure 4).
Sf3b1K700E/+ mice develop progressive normocytic anemia without ring sideroblasts. (a) Sf3b1 alleles in study mice. The conditional (pre-Cre) allele contains a floxed splice trap cassette inserted within intron 11 of the gene. This begins with a splice acceptor site (SA) splicing in-frame into exons 12–25 of a partially codon-optimized (c/o) Sf3b1 cDNA and ending with a polyadenylation signal (pA). This allele therefore expresses a chimeric mRNA, which is translated into the wild-type (WT) SF3B1 protein. Downstream of this, the endogenous exon 15 (red) was mutagenized to encode the K700E mutation. After Cre-mediated excision of the trap cassette, the recombined allele (post-Cre) expresses Sf3b1-K700E. (b) Serial blood counts highlight a progressive normocytic anemia in Sf3b1K700E/+ compared to age-matched WT mice. Platelet (PLT) and leukocyte counts (WBC) were not different between mutant and WT mice. (c) Examination of Sf3b1K700E/+ bone marrow cytospins showed no significant evidence of dysplasia (upper panel) and Perl staining demonstrated macrophage iron (arrows), but no ring sideroblasts (lower panel). (d) In keeping with the cytological findings, histological examination did not show significant dysplasia (upper panel), although Perl staining did reveal increased iron deposits in mutant bone marrow macrophages (lower panel). Hb, hemoglobin concentration; MCV, mean corpuscular volume.
To activate Sf3b1K700E in hematopoietic stem cells, we crossed Sf3b1flox-K700E/+ and Mx1-Cre23 mice, treated 20 double transgenic and 20 control (wild type and Sf3b1flox-K700E/+; Mx1-Cre-, henceforth referred as WT) 4- to 6-week-old mice with polyinosinic-polycytidylic acid (pIpC) and monitored the animals longitudinally. Monthly blood counts from mutant mice (henceforth referred to as Sf3b1K700E/+) showed a progressive normocytic anemia compared to wild type, but no significant differences in white blood cell (WBC) or platelet (PLT) counts (Figure 1b). Cytological examination of the bone marrow of Sf3b1K700E/+ mice showed no significant dysplasia and no ring sideroblasts (Figure 1c). Bone marrow (BM) histology did not reveal any significant morphological differences between WT and Sf3b1K700E/+ mice, but did show a moderate overall increase in iron deposits in the latter (Figure 1d). At a median follow-up of 83 weeks, mutant animals did not show altered survival or increased signs of illness when compared to WT controls (Supplementary Figure 5a).
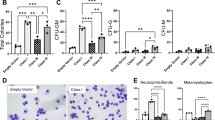
To investigate the consequences of Sf3b1K700E expression on hematopoiesis in greater detail, we analyzed equal numbers of WT and mutant bone marrow cells harvested 4 weeks after pIpC injection by flow cytometry. We found that Sf3b1K700E/+ mice showed a significant decrease in the number of phenotypically defined hematopoietic stem cells (HSCs), but no change in the size of the LMPP, GMP, CMP and MEP progenitor compartments compared to WT littermates (Figure 2a). By contrast, Sf3b1K700E/+ animals showed a small but significant increase in the number of Gr1+/Mac1+ double-positive bone marrow cells (Supplementary Figure 5b), and in the erythroid compartment there were no differences in the frequency of Ter119+/CD71+ early erythroid cells, but a relative decrease of the more mature Ter119+/CD71low/FSClow population24 (Figure 2b), indicative of impaired terminal erythroid differentiation similar to that observed in human MDS.25
Effects of mutant Sf3b1K700E/+ on hemopoietic stem and progenitor cells. (a) Flow cytometric analysis of bone marrow cells from Sf3b1K700E/+ mice showed a significant decrease of phenotypically defined HSCs compared to WT animals, but no differences in LMPP, GMP, CMP or MEP progenitor numbers (n = 4). (b) Early erythroid cells (Ter119+/CD71+) were unchanged in number, but there was a reduction in mature Ter119low/CD71+ erythoid cells in Sf3b1K700E/+ animals. (c) Competitive transplantation of bone marrow lin− cells from WT (CD45.1) and Sf3b1K700E/+ (CD45.2) into young (2 months) or old (1 year) syngeneic (C57BL/6) recipients (n=4 per group). Results shows that, after good initial engraftment (1 month), Sf3b1K700E/+ (CD45.2) progenitor cells were steadily outcompeted by co-transplanted WT (CD45.1) cells. Bars in (a, b) and datapoints in (c) show mean±standard error of the mean.
Next, as SF3B1-K700E behaves like an initiating mutation in MDS7 and can also drive clonal hematopoiesis in elderly individuals,8 we wanted to investigate if it confers a fitness advantage to murine HSCs. To test this, we performed competitive transplantation assays in lethally irradiated syngeneic (C57BL/6) recipient mice. These experiments were performed in both young (2 months) and old (12 months) animals to investigate the possibility that age-related changes in recipient mice may influence the repopulating fitness of Sf3b1K700E/+ mutant stem cells.26 Experimental mice were transplanted with an equal number (2.5 × 105) of bone marrow lineage negative (lin−) progenitor cells from WT (CD45.1) and Sf3b1K700E/+ (CD45.2) mice, as well as 2.5 × 105 whole bone marrow (rescue) cells from Sf3b1+/+ (CD45.1/CD45.2) animals. The control group was transplanted with rescue cells from whole bone marrow (CD45.1/CD45.2 = 1 × 106). We found that although Sf3b1K700E/+ cells did show good engraftment at 1 month post transplantation, they displayed a significant decline in their white blood cell progeny after 4 months compared to WT cells. This was true for both young and old recipients (Figure 2c). Similar results were obtained using a different Sf3b1K700E/+ donor mouse.
Since SF3B1 mutations affect pre-mRNA splicing and consequently gene expression,13 we performed mRNA sequencing from whole bone marrow (BM) and lin− progenitors of Sf3b1K700E/+ and WT animals. Firstly, we observed that in Sf3b1K700E/+ mice transcript levels of the mutant (K700E) and wild-type Sf3b1 mRNAs were very similar (Supplementary Table 3), confirming that expression from the targeted allele was not altered by genetic modification of the locus. We next sought to identify aberrant splicing events associated with Sf3b1-K700E using splice site analysis of RNAseq data from Sf3b1K700E/+ and WT BM cells as we described before13 (also see Materials and Methods). Compared to WT we identified 719 aberrant events in lin− and 293 in whole BM cells from Sf3b1K700E/+ mice. The dominant type of aberrant splicing event was increased usage of cryptic 3′ splice sites (3′ ss) (Figure 3a), the majority of which were novel, that is, not reported in current transcriptome annotations. Aberrant splice sites were located within a narrow window of 15–24 nucleotides (nt) upstream from the canonical 3′ ss (Figure 3b) and associated with sequence features, including a shorter polypyrimidine tract and an enrichment of adenines at positions -8 to -18 upstream of the cryptic 3′ ss (Figures 3c and d), as described for human cancers with SF3B1 mutations.13 Since SF3B1 is known to play a major role in U2 snRNP recruitment to the splicing branchpoint (BP), we performed lariat sequencing to identify BPs in aberrantly spliced mouse mRNAs. One example we identified was the Ugdh mRNA, for which the BP was located 16 nt upstream of the aberrant AG splice acceptor site that notably was not conserved in the human genome (Figure 3e). The Get4 gene showed similar features (Supplementary Figure 6). These observations demonstrate that, as shown recently for human cancers,11 there is a distinct usage of cryptic BPs in cells expressing mutant SF3B1 in our mouse model.
Reconstruction of annotated transcripts that used aberrant 3′ splice sites in the lin− RNAseq dataset revealed that approximately 42% of affected genes gain a premature termination codon in all isoforms, which is predicted to result in mRNA degradation through the nonsense-mediated decay (NMD) pathway. Genes for which NMD was predicted (n = 190) showed significantly lower mRNA expression levels than those for which NMD was not predicted (n = 225) (P = 3.92 × 10−13, Kruskal H test, Figure 3f). Furthermore, looking at the transcriptome as a whole, we found that most (408 of 511) differentially expressed genes in lin− cells were downregulated (discussed below). These findings demonstrate that Sf3b1-K700E is associated with abnormal splicing in hematopoietic stem/progenitor cells, primarily through the use of aberrant 3′-ss that are thought to result in NMD, and this is associated with significant downregulation of a large number of mRNAs.
Unbiased gene set enrichment analysis (GSEA) of the genes differentially expressed in Sf3b1K700E/+ revealed an enrichment of genes involved in RNA processing, splicing and transcription. Interestingly, similar enrichment was observed in human MDS11 (Supplementary Figure 7).
Broad transcriptomic changes were observed in human cancers with SF3B1 mutations including MDS,11, 27 and we recently described the aberrant splicing patterns in such cancers.13 Here we perform a similar analysis on RNAseq data from Sf3b1K700E/+ mice and report that for aberrant 3′ splice sites with upstream cryptic AGs, splicing abnormalities are strikingly similar to those seen in human cancer samples, including the aberrant splice site consensus sequence (Figure 4a) and distance of aberrant 3′ splice sites from the canonical ones (Figure 4b). However, despite these striking similarities, there was minimal overlap between aberrantly spliced genes in mouse vs human cells. In fact, comparing the two datasets we found that only ~5% (17/360) of the aberrantly spliced genes in mouse Sf3b1K700E/+ lin− cells were also aberrantly spliced in human samples (Figure 4c). To look at this more closely, we compared the nucleotide sequences at the location of the aberrant 3′ splice sites identified in human SF3B1-mutant cancers,11, 13 including MDS, and those identified in our Sf3b1K700E/+ mouse BM and lin− cells with the equivalent positions in the genome of the other species, examining the conservation of the invariant AG dinucleotides at the splice sites and that of the upstream 35nt. We first examined genes thought to have a role in the formation of ring sideroblasts in human SF3B1-mutant MDS, namely TMEM14C25 and ABCB7,28, 29 for which aberrant 3′ splicing was observed in humans, but not in Sf3b1K700E/+ mice. The human TMEM14C aberrant splice site AG was not conserved in mice, while for ABCB7, although the AG dinucleotide was conserved, it was located 2 nt upstream and was preceded by a poorly conserved sequence that deviated significantly from the aberrant splicing consensus (Figure 4d). We then looked at the 360 canonical junctions with aberrant upstream 3′ splice sites in Sf3b1K700E/+ lin− cells and found that 268 were conserved in the human genome. Of the 268 human junctions only 125 contained both a potential cryptic AG in the range preferred by SF3B1/Sf3b1 mutants (-5 to -20nt) and a potential branchpoint adenosine at -8 to -18 from the cryptic AG, indicating that less than 35% of mouse junctions with aberrant 3′ splice sites had the hypothetical capacity for aberrant splicing in human cells. Of these 125 potential aberrant junctions, only 53 (14.7% of total) were found to be spliced in any human MDS sample, indicating that the limited conservation between mouse and human intronic sequences is responsible for the differences in misspliced mRNAs in the two species. We also noted that both Tmem14c and Abcb7 as well as the Alas2 (heme biosynthesis) and Slc25a37 (mitochondrial iron importer) genes that are downregulated in human SF3B1-mutant MDS11 did not display altered expression in Sf3b1K700E/+ lin− or whole BM cells (Figure 4e).
Discussion
In order to understand the molecular and phenotypic consequences of SF3B1 mutations on hematopoiesis and their role in the pathogenesis of clonal blood disorders, we generated a knock-in conditional mouse model of SF3B1-K700E, the most common mutation in human MDS. Our conditional allele preserved wild-type Sf3b1 expression, thus avoiding any confounding effects of reduced Sf3b1 levels on hematopoiesis11, 30 or other developmental effects. Conditional activation by Mx1-Cre of the Sf3b1-K700E mutation in Sf3b1flox−K700E/+ mice then enabled direct comparison to wild-type (WT) isogenic hematopoietic progenitors at the molecular and phenotypic level. At the molecular level, we report that the splicing defect seen in Sf3b1K700E/+ hematopoietic progenitors closely mimicked that described in human MDS, in sharing a strikingly similar splice-site consensus sequence and distance from the aberrant splice site AG. Furthermore, as was recently described for human cancer samples,31, 32 we show that aberrant BP recognition is central to the disrupted function of mutant SF3B1 in our mice. However, despite the closely similar properties of human and mouse SF3B1 mutants, we found that misspliced transcripts differed significantly between mouse and human as a result of the relatively poor interspecies conservation of intronic sequences able to function as aberrant splice sites. Interestingly, a recent study describing a murine model of U2AF1-S34F, another MDS-associated splicing gene mutation associated with aberrant 3′ splice acceptor site recognition, also showed limited mouse–human overlap of mispliced transcripts.33 The mouse–human overlap appeared greater for a mouse model of SRSF2 mutations (SRSF2-P95H), another commonly mutated gene in human MDS, possibly because the splicing aberration involves alternative exonic sequences, which are more likely than intronic ones to be conserved.34
With regard to erythropoiesis, Sf3b1K700E/+ mice showed impaired terminal erythroid differentiation, associated with progressive normocytic anemia without ring sideroblasts. In human SF3B1-mutant MDS, ring sideroblasts are thought to arise as a result of aberrant splicing of key genes involved in heme biosynthesis, such as ABCB7, TMEM14C, ALAS2 and SLC25A37.11, 25, 30, 35 In keeping with the mouse–human differences in target transcripts, we did not find the splicing or expression levels of any of these genes to be noticeably disrupted by Sf3b1-K700E, and, consistent with this, did not identify ring sideroblasts in the bone marrow of mutant mice. Of note, ring sideroblasts were not observed in single-gene knock-out mice for either Alas236 or Abcb7,37 suggesting that sideroblast formation may require simultaneous reduction in the levels of multiple proteins involved in both heme biosynthesis (ALAS2, TMEM14C) and mitochondrial iron transport (ABCB7 and SLC25A37), and may therefore be difficult to replicate in single-gene targeting models. In this light, the development of anemia in Sf3b1K700E/+ mice may be at least in part due to globally disrupted splicing rather than effects on particular genes. Aberrant splicing leads to the introduction of premature termination codons, which can in turn lead to the generation of truncated proteins with potential toxic effects on cells, leading in this instance to anemia and impaired iron utilization manifested as excess hemosiderin deposition in the bone marrow of Sf3b1K700E/+ mice. An alternative or additional explanation may reside in the fact that both mouse and human cells displayed aberrant expression of genes involved in splicing and RNA processing itself (Supplementary Figure 7), which may be a response to, rather than a result of, globally disrupted splicing. In fact there is evidence that when their intracellular concentrations become too high, some splicing factors regulate their own expression by targeting their mRNAs for NMD.38
Beyond erythropoiesis, Sf3b1K700E/+ mice had reduced numbers of HSCs and displayed a myeloid cell bias. Furthermore, investigation of the self-renewal potential of Sf3b1K700E/+ HSCs as determined by their repopulating ability in competitive transplantation assays into either young or old recipient mice revealed a fitness disadvantage of mutant over wild-type HSCs. This contrasts with observations that mutant SF3B1 can drive clonal hematopoiesis and can even be the sole identifiable driver mutation in human MDS.7, 9 However, observations that SF3B1 mutations appear to selectively impart a clonal advantage on HSCs of elderly, but not young, individuals suggest that the phenomenon is context dependent even in humans.26 Furthermore, a lack of clonal HSC advantage was a feature of other mouse models of MDS-associated mutations, including ASXL1,39 U2AF133 and SRSF2,34 all of which can also drive clonal hematopoiesis in humans,40, 41 proposing that factors other than somatic mutations may be operative in the development of MDS.26
The fact that disruption of a process as central to cellular function as RNA splicing can have detrimental effects on cells is unsurprising and further augments the conundrum of how particular spliceosome gene mutations can instead impart a survival/fitness benefit upon human HSCs, albeit in a context-dependent manner.8, 26 This remains the least-well-understood feature of spliceosome gene mutations, and our data propose that the different phenotypic effects of spliceosome gene mutations in MDS (e.g. anemia, ring sideroblasts, clonal expansion) may have distinct molecular causes. Future studies aimed at deciphering the mechanisms underlying these phenotypes should exploit both the similarities and the differences in the transcriptional consequences of these mutations between mice and humans, as a means of identifying their respective etiologies.
References
Tefferi A, Vardiman JW . Myelodysplastic syndromes. N Engl J Med 2009; 361: 1872–1885.
Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011; 365: 1384–1395.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69.
Vardiman JW . The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: an overview with emphasis on the myeloid neoplasms. Chem Biol Interact 2010; 184 16–20.
Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011; 118: 6239–6246.
Mian SA, Rouault-Pierre K, Smith AE, Seidl T, Pizzitola I, Kizilors A et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat Commun 2015; 6: 10004.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122: 3616–3627, quiz 99.
McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 2015; 10: 1239–1245.
Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014; 28: 241–247.
Scotti MM, Swanson MS . RNA mis-splicing in disease. Nat Rev Genet 2016; 17: 19–32.
Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015; 29: 1092–1103.
Gerstung M, Pellagatti A, Malcovati L, Giagounidis A, Porta MG, Jadersten M et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun 2015; 6: 5901.
Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3' splice site selection through use of a different branch point. Cell Rep 2015; 13: 1033–1045.
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011; 474: 337–342.
Liu P, Jenkins NA, Copeland NG . A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003; 13: 476–484.
Li J, Spensberger D, Ahn JS, Anand S, Beer PA, Ghevaert C et al. JAK2 V617F impairs hematopoietic stem cell function in a conditional knock-in mouse model of JAK2 V617F-positive essential thrombocythemia. Blood 2010; 116: 1528–1538.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3.
Patro R, Mount SM, Kingsford C . Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 2014; 32: 462–464.
Robinson MD, McCarthy DJ, Smyth GK . edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140.
Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 2009; 6: 493–495.
Farley FW, Soriano P, Steffen LS, Dymecki SM . Widespread recombinase expression using FLPeR (flipper) mice. Genesis 2000; 28: 106–110.
Kuhn R, Schwenk F, Aguet M, Rajewsky K . Inducible gene targeting in mice. Science 1995; 269: 1427–1429.
Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N . Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA 2009; 106: 17413–17418.
Conte S, Katayama S, Vesterlund L, Karimi M, Dimitriou M, Jansson M et al. Aberrant splicing of genes involved in haemoglobin synthesis and impaired terminal erythroid maturation in SF3B1 mutated refractory anaemia with ring sideroblasts. Br J Haematol. 2015; 171: 478–490.
McKerrell T, Vassiliou GS . Aging as a driver of leukemogenesis. Sci Transl Med 2015; 7: 306fs38.
Gentien D, Kosmider O, Nguyen-Khac F, Albaud B, Rapinat A, Dumont AG et al. A common alternative splicing signature is associated with SF3B1 mutations in malignancies from different cell lineages. Leukemia 2014; 28: 1355–1357.
Nikpour M, Scharenberg C, Liu A, Conte S, Karimi M, Mortera-Blanco T et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 2013; 27: 889–896.
Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 2016; e-pub ahead of print 17 June 2016 doi:10.1038/leu.2016.149.
Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood 2012; 120: 3173–3186.
Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 2016; 7: 10615.
DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, Jepsen K et al. Transcriptome sequencing reveals potential mechanism of cryptic 3' splice site selection in SF3B1-mutated cancers. PLoS Comput Biol 2015; 11: e1004105.
Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell 2015; 27: 631–643.
Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015; 27: 617–630.
Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006; 440: 96–100.
Harigae H, Nakajima O, Suwabe N, Yokoyama H, Furuyama K, Sasaki T et al. Aberrant iron accumulation and oxidized status of erythroid-specific delta-aminolevulinate synthase (ALAS2)-deficient definitive erythroblasts. Blood 2003; 101: 1188–1193.
Pondarre C, Campagna DR, Antiochos B, Sikorski L, Mulhern H, Fleming MD . Abcb7 the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 2007; 109: 3567–3569.
McGlincy NJ, Smith CW . Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 2008; 33: 385–393.
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 2013; 210: 2641–2659.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371: 2477–2487.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498.
Acknowledgements
This work was funded by the Wellcome Trust Sanger Institute (WT098051). AM is funded by the Kay Kendall Leukaemia Fund (KKL634). GSV is funded by a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA) and work in his laboratory is also funded by the Wellcome Trust Sanger Institute (WT098051), Bloodwise and the Kay Kendal Leukaemia Fund. GSV is a consultant for and holds stock in Kymab Ltd, and receives an educational grant from Celgene. NB is funded by the Associazione Italiana Ricerca sul Cancro (MFAG AIRC grant n. 17658). MS, AA and SB are employees of H3 Biomedicine. We thank Frances Law for her help with ES targeting and cell culture. We thank Peggy Goodell for helpful discussions about this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Mupo, A., Seiler, M., Sathiaseelan, V. et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 31, 720–727 (2017). https://doi.org/10.1038/leu.2016.251
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.251
This article is cited by
-
The evolution of preclinical models for myelodysplastic neoplasms
Leukemia (2024)
-
Bone marrow inflammation in haematological malignancies
Nature Reviews Immunology (2024)
-
Exploring the mechanistic link between SF3B1 mutation and ring sideroblast formation in myelodysplastic syndrome
Scientific Reports (2022)
-
Alternative splicing and cancer: a systematic review
Signal Transduction and Targeted Therapy (2021)
-
Complex landscape of alternative splicing in myeloid neoplasms
Leukemia (2021)