Abstract
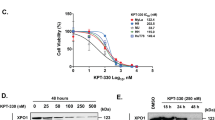
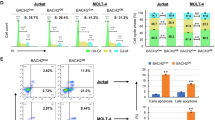
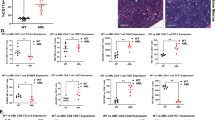
The bridging integrator 1 (BIN1) tumor suppressor encodes multiple alternatively spliced isoforms implicated in DNA repair, cell-cycle control, apoptosis and membrane dynamics. BIN1 attenuation has been reported in several solid tumors; however, the role of BIN1 in lymphomagenesis remains unexplored. We recently demonstrated that BIN1 transcript levels are significantly downregulated in CD4+CD7− Sezary cells from patients with Sezary syndrome (SS), a subtype of cutaneous T-cell lymphoma (CTCL). We have now demonstrated that restored BIN1 expression in CTCL cells leads to a significant reduction in cell proliferation, an increase in spontaneous and Fas/Fas ligand (Fas/FasL)-induced apoptosis in vitro and inhibition of tumorigenic activity of CTCL cells in vivo. Interestingly, restoration of BIN1 expression in CTCL cells downregulates the expression of c-FLIP, an important inhibitor of Fas/FasL-mediated apoptosis, and activates the caspase cascade; these phenotypes can be rescued by knockdown of BIN1. Importantly, significantly reduced BIN1 expression and increased c-FLIP expression are observed in primary CTCL patient samples, and high BIN1 and low c-FLIP mRNA levels correlate with better survival rate in SS patients. These results indicate that BIN1 regulates Fas/FasL-mediated apoptosis through c-FLIP and that BIN1 deficiency may have an important role in CTCL pathogenesis by causing apoptosis resistance. Thus BIN1 and c-FLIP represent potential therapeutic targets in CTCL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Li JY, Horwitz S, Moskowitz A, Myskowski PL, Pulitzer M, Querfeld C . Management of cutaneous T cell lymphoma: new and emerging targets and treatment options. Cancer Manag Res 2012; 4: 75–89.
Klemke CD, Goerdt S, Schrama D, Becker JC . New insights into the molecular biology and targeted therapy of cutaneous T-cell lymphomas. J Dtsch Dermatol Ges 2006; 4: 395–406.
Hwang ST, Janik JE, Jaffe ES, Wilson WH . Mycosis fungoides and Sezary syndrome. Lancet 2008; 371: 945–957.
Booken N, Gratchev A, Utikal J, Weiss C, Yu X, Qadoumi M et al. Sezary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia 2008; 22: 393–399.
Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med 2003; 197: 1477–1488.
Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007; 110: 1713–1722.
Dulmage BO, Geskin LJ . Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol 2013; 169: 1188–1197.
Wang Y, Su M, Zhou LL, Tu P, Zhang X, Jiang X et al. Deficiency of SATB1 expression in Sezary cells causes apoptosis resistance by regulating FasL/CD95L transcription. Blood 2011; 117: 3826–3835.
Mao X, Orchard G, Mitchell TJ, Oyama N, Russell-Jones R, Vermeer MH et al. A genomic and expression study of AP-1 in primary cutaneous T-cell lymphoma: evidence for dysregulated expression of JUNB and JUND in MF and SS. J Cutan Pathol 2008; 35: 899–910.
Guenova E, Hoetzenecker W, Rozati S, Levesque MP, Dummer R, Cozzio A . Novel therapies for cutaneous T-cell lymphoma: what does the future hold? Expert Opin Investig Drugs 2014; 23: 457–467.
van Doorn R, van Kester MS, Dijkman R, Vermeer MH, Mulder AA, Szuhai K et al. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood 2009; 113: 127–136.
Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011; 118: 5891–5900.
Contassot E, Kerl K, Roques S, Shane R, Gaide O, Dupuis M et al. Resistance to FasL and tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in Sezary syndrome T-cells associated with impaired death receptor and FLICE-inhibitory protein expression. Blood 2008; 111: 4780–4787.
Braun FK, Fecker LF, Schwarz C, Walden P, Assaf C, Durkop H et al. Blockade of death receptor-mediated pathways early in the signaling cascade coincides with distinct apoptosis resistance in cutaneous T-cell lymphoma cells. J Invest Dermatol 2007; 127: 2425–2437.
Wu J, Nihal M, Siddiqui J, Vonderheid EC, Wood GS . Low FAS/CD95 expression by CTCL correlates with reduced sensitivity to apoptosis that can be restored by FAS upregulation. J Invest Dermatol 2009; 129: 1165–1173.
Wu J, Siddiqui J, Nihal M, Vonderheid EC, Wood GS . Structural alterations of the FAS gene in cutaneous T-cell lymphoma (CTCL). Arch Biochem Biophys 2011; 508: 185–191.
Valente G, Manfroi F, Peracchio C, Nicotra G, Castino R, Nicosia G et al. cFLIP expression correlates with tumour progression and patient outcome in non-Hodgkin lymphomas of low grade of malignancy. Br J Haematol 2006; 132: 560–570.
Dummer R, Michie SA, Kell D, Gould JW, Haeffner AC, Smoller BR et al. Expression of bcl-2 protein and Ki-67 nuclear proliferation antigen in benign and malignant cutaneous T-cell infiltrates. J Cutan Pathol 1995; 22: 11–17.
van Doorn R, Dijkman R, Vermeer MH, Starink TM, Willemze R, Tensen CP . A novel splice variant of the Fas gene in patients with cutaneous T-cell lymphoma. Cancer Res 2002; 62: 5389–5392.
Ringrose A, Zhou Y, Pang E, Zhou L, Lin AE, Sheng G et al. Evidence for an oncogenic role of AHI-1 in Sezary syndrome, a leukemic variant of human cutaneous T-cell lymphomas. Leukemia 2006; 20: 1593–1601.
Kennah E, Ringrose A, Zhou LL, Esmailzadeh S, Qian H, Su MW et al. Identification of tyrosine kinase, HCK, and tumor suppressor, BIN1, as potential mediators of AHI-1 oncogene in primary and transformed CTCL cells. Blood 2009; 113: 4646–4655.
Esmailzadeh S, Jiang X . AHI-1: a novel signaling protein and potential therapeutic target in human leukemia and brain disorders. Oncotarget 2011; 2: 918–934.
Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC . BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet 1996; 14: 69–77.
Wechsler-Reya RJ, Elliott KJ, Prendergast GC . A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol Cell Biol 1998; 18: 566–575.
Elliott K, Ge K, Du W, Prendergast GC . The c-Myc-interacting adaptor protein Bin1 activates a caspase-independent cell death program. Oncogene 2000; 19: 4669–4684.
DuHadaway JB, Lynch FJ, Brisbay S, Bueso-Ramos C, Troncoso P, McDonnell T et al. Immunohistochemical analysis of Bin1/Amphiphysin II in human tissues: diverse sites of nuclear expression and losses in prostate cancer. J Cell Biochem 2003; 88: 635–642.
Cassimere EK, Pyndiah S, Sakamuro D . The c-MYC-interacting proapoptotic tumor suppressor BIN1 is a transcriptional target for E2F1 in response to DNA damage. Cell Death Differ 2009; 16: 1641–1653.
Kinney EL, Tanida S, Rodrigue AA, Johnson JK, Tompkins VS, Sakamuro D . Adenovirus E1A oncoprotein liberates c-Myc activity to promote cell proliferation through abating Bin1 expression via an Rb/E2F1-dependent mechanism. J Cell Physiol 2008; 216: 621–631.
Prokic I, Cowling BS, Laporte J . Amphiphysin 2 (BIN1) in physiology and diseases. J Mol Med (Berl) 2014; 124: 976–978.
Ge K, Duhadaway J, Sakamuro D, Wechsler-Reya R, Reynolds C, Prendergast GC . Losses of the tumor suppressor BIN1 in breast carcinoma are frequent and reflect deficits in programmed cell death capacity. Int J Cancer 2000; 85: 376–383.
Tajiri T, Liu X, Thompson PM, Tanaka S, Suita S, Zhao H et al. Expression of a MYCN-interacting isoform of the tumor suppressor BIN1 is reduced in neuroblastomas with unfavorable biological features. Clin Cancer Res 2003; 9: 3345–3355.
Chang MY, Boulden J, Katz JB, Wang L, Meyer TJ, Soler AP et al. Bin1 ablation increases susceptibility to cancer during aging, particularly lung cancer. Cancer Res 2007; 67: 7605–7612.
Prendergast GC, Muller AJ, Ramalingam A, Chang MY . BAR the door: cancer suppression by amphiphysin-like genes. Biochim Biophys Acta 2009; 1795: 25–36.
Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast GC . Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci USA 1999; 96: 9689–9694.
Ghaneie A, Zemba-Palko V, Itoh H, Itoh K, Sakamuro D, Nakamura S et al. Bin1 attenuation in breast cancer is correlated to nodal metastasis and reduced survival. Cancer Biol Ther 2007; 6: 192–194.
Ge K, Minhas F, Duhadaway J, Mao NC, Wilson D, Buccafusca R et al. Loss of heterozygosity and tumor suppressor activity of Bin1 in prostate carcinoma. Int J Cancer 2000; 86: 155–161.
Galderisi U, Di Bernardo G, Cipollaro M, Jori FP, Piegari E, Cascino A et al. Induction of apoptosis and differentiation in neuroblastoma and astrocytoma cells by the overexpression of Bin1, a novel Myc interacting protein. J Cell Biochem 1999; 74: 313–322.
Elliott K, Sakamuro D, Basu A, Du W, Wunner W, Staller P et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene 1999; 18: 3564–3573.
Pineda-Lucena A, Ho CS, Mao DY, Sheng Y, Laister RC, Muhandiram R et al. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J Mol Biol 2005; 351: 182–194.
Zhou LL, Zhao Y, Ringrose A, DeGeer D, Kennah E, Lin AE et al. AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells. J Exp Med 2008; 205: 2657–2671.
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998; 72: 8463–8471.
Chen M, Gallipoli P, DeGeer D, Sloma I, Forrest DL, Chan M et al. Targeting primitive chronic myeloid leukemia cells by effective inhibition of a new AHI-1-BCR-ABL-JAK2 complex. J Natl Cancer Inst 2013; 105: 405–423.
Jiang X, Saw KM, Eaves A, Eaves C . Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst 2007; 99: 680–693.
Euhus DM, Hudd C, LaRegina MC, Johnson FE . Tumor measurement in the nude mouse. J Surg Oncol 1986; 31: 229–234.
Tomayko MM, Reynolds CP . Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 1989; 24: 148–154.
Camp RL, Dolled-Filhart M, Rimm DL . X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10: 7252–7259.
Wechsler-Reya R, Sakamuro D, Zhang J, Duhadaway J, Prendergast GC . Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J Biol Chem 1997; 272: 31453–31458.
Diamandidou E, Cohen PR, Kurzrock R . Mycosis fungoides and Sezary syndrome. Blood 1996; 88: 2385–2409.
Siegel RS, Pandolfino T, Guitart J, Rosen S, Kuzel TM . Primary cutaneous T-cell lymphoma: review and current concepts. J Clin Oncol 2000; 18: 2908–2925.
Kuzel TM . Systemic chemotherapy for the treatment of mycosis fungoides and Sezary syndrome. Dermatol Ther 2003; 16: 355–361.
Valnet-Rabier MB, Challier B, Thiebault S, Angonin R, Margueritte G, Mougin C et al. c-Flip protein expression in Burkitt's lymphomas is associated with a poor clinical outcome. Br J Haematol 2005; 128: 767–773.
El-Gazzar A, Wittinger M, Perco P, Anees M, Horvat R, Mikulits W et al. The role of c-FLIP(L) in ovarian cancer: chaperoning tumor cells from immunosurveillance and increasing their invasive potential. Gynecol Oncol 2010; 117: 451–459.
Day TW, Huang S, Safa AR . c-FLIP knockdown induces ligand-independent DR5-, FADD-, caspase-8-, and caspase-9-dependent apoptosis in breast cancer cells. Biochem Pharmacol 2008; 76: 1694–1704.
Haag C, Stadel D, Zhou S, Bachem MG, Moller P, Debatin KM et al. Identification of c-FLIP(L) and c-FLIP(S) as critical regulators of death receptor-induced apoptosis in pancreatic cancer cells. Gut 2011; 60: 225–237.
Yang BF, Xiao C, Li H, Yang SJ . Resistance to Fas-mediated apoptosis in malignant tumours is rescued by KN-93 and cisplatin via downregulation of c-FLIP expression and phosphorylation. Clin Exp Pharmacol Physiol 2007; 34: 1245–1251.
Logan AE, Wilson TR, Fenning C, Cummins R, Kay E, Johnston PG et al. In vitro and in vivo characterisation of a novel c-FLIP-targeted antisense phosphorothioate oligonucleotide. Apoptosis 2010; 15: 1435–1443.
Day TW, Najafi F, Wu CH, Safa AR . Cellular FLICE-like inhibitory protein (c-FLIP): a novel target for Taxol-induced apoptosis. Biochem Pharmacol 2006; 71: 1551–1561.
Safa AR, Pollok KE . Targeting the anti-apoptotic protein c-FLIP for cancer therapy. Cancers (Basel) 2011; 3: 1639–1671.
Safa AR, Day TW, Wu CH . Cellular FLICE-like inhibitory protein (C-FLIP): a novel target for cancer therapy. Curr Cancer Drug Targets 2008; 8: 37–46.
Acknowledgements
We thank Terry Fox Laboratory FACS Facility for cell sorting; Dr George Prendergast for BIN1 cDNA plasmids and BIN1 (12A) antibody; Dr Linda Penn for BIN1 12A plasmid; Helena Wang and Josephine Leung for excellent technical assistance; and STEMCELL Technologies for cell culture reagents. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and Canadian Dermatology Foundation (to YZ), the Leukemia & Lymphoma Society of Canada and in part by CIHR, the Canadian Cancer Society and the Cancer Research Society (to XJ). SE and YH are recipients of CIHR Skin Research Training Center Scholarships. YH is a recipient of the Vanier Canada Graduate Scholarship.
Author Contributions
SE, YZ and XJ developed the concept and designed the experiments; SE performed most experiments and analyzed data; YH generated the Kaplan–Meier curve and assisted with q-RT-PCR on primary SS samples; and MS extracted RNA from MF patient samples. SE and XJ wrote the manuscript and the other authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Esmailzadeh, S., Huang, Y., Su, MW. et al. BIN1 tumor suppressor regulates Fas/Fas ligand–mediated apoptosis through c-FLIP in cutaneous T-cell lymphoma. Leukemia 29, 1402–1413 (2015). https://doi.org/10.1038/leu.2015.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.9
This article is cited by
-
The Role of Changes in the Expression of Inflammation-Associated Genes in Cerebral Small Vessel Disease with Cognitive Impairments
Neuroscience and Behavioral Physiology (2024)
-
BIN1 in cancer: biomarker and therapeutic target
Journal of Cancer Research and Clinical Oncology (2023)
-
The Prognostic Significance of the BIN1 and CCND2 Gene in Adult Patients with Acute Myeloid Leukemia
Indian Journal of Hematology and Blood Transfusion (2022)
-
Targeting CASP8 and FADD-like apoptosis regulator ameliorates nonalcoholic steatohepatitis in mice and nonhuman primates
Nature Medicine (2017)
-
Malignant inflammation in cutaneous T‐cell lymphoma—a hostile takeover
Seminars in Immunopathology (2017)