Abstract
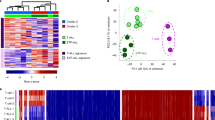
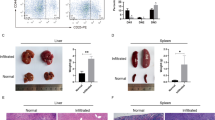
The TLX1 transcription factor is critically involved in the multi-step pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL) and often cooperates with NOTCH1 activation during malignant T-cell transformation. However, the exact molecular mechanism by which these T-cell specific oncogenes cooperate during transformation remains to be established. Here, we used chromatin immunoprecipitation followed by sequencing to establish the genome-wide binding pattern of TLX1 in human T-ALL. This integrative genomics approach showed that ectopic TLX1 expression drives repression of T cell-specific enhancers and mediates an unexpected transcriptional antagonism with NOTCH1 at critical target genes, including IL7R and NOTCH3. These phenomena coordinately trigger a TLX1-driven pre-leukemic phenotype in human thymic precursor cells, reminiscent of the thymus regression observed in murine TLX1 tumor models, and create a strong genetic pressure for acquiring activating NOTCH1 mutations as a prerequisite for full leukemic transformation. In conclusion, our results uncover a functional antagonism between cooperative oncogenes during the earliest phases of tumor development and provide novel insights in the multi-step pathogenesis of TLX1-driven human leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ferrando AA, Neuberg DS, Dodge RK, Paietta E, Larson RA, Wiernik PH et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet 2004; 363: 535–536.
Hawley RG, Fong AZ, Reis MD, Zhang N, Lu M, Hawley TS . Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res 1997; 57: 337–345.
Hawley RG, Fong AZ, Lu M, Hawley TS . The HOX11 homeobox-containing gene of human leukemia immortalizes murine hematopoietic precursors. Oncogene 1994; 9: 1–12.
De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med 2010; 16: 1321–1327.
Rakowski LA, Lehotzky EA, Chiang MY . Transient responses to NOTCH and TLX1/HOX11 inhibition in T-cell acute lymphoblastic leukemia/lymphoma. PLoS ONE 2011; 6: e16761.
Owens BM, Hawley TS, Spain LM, Kerkel KA, Hawley RG . TLX1/HOX11-mediated disruption of primary thymocyte differentiation prior to the CD4+CD8+ double-positive stage. Br J Haematol 2006; 132: 216–229.
Dadi S, Le Noir S, Payet-Bornet D, Lhermitte L, Zacarias-Cabeza J, Bergeron J et al. TLX homeodomain oncogenes mediate T cell maturation arrest in T-ALL via interaction with ETS1 and suppression of TCRalpha gene expression. Cancer Cell 2012; 21: 563–576.
Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci USA 2014; 111: 705–710.
Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA 2011; 108: 14908–14913.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood 2009; 113: 3918–3924.
Clappier E, Gerby B, Sigaux F, Delord M, Touzri F, Hernandez L et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med 2011; 208: 653–661.
Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 2013; 41: D246–D251.
Lee TI, Johnstone SE, Young RA . Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 2006; 1: 729–748.
Langmead B, Trapnell C, Pop M, Salzberg SL . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25.
Feng J, Liu T, Zhang Y . Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics 2011; Chapter 2: Unit 2.14..
Quinlan AR, Hall IM . BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26: 841–842.
Machanick P, Bailey TL . MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011; 27: 1696–1697.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589.
Van de Walle I, Waegemans E, De Medts J, De Smet G, De Smedt M, Snauwaert S et al. Specific Notch receptor-ligand interactions control human TCR-alphabeta/gammadelta development by inducing differential Notch signal strength. J Exp Med 2013; 210: 683–697.
Van de Walle I, De Smet G, Gartner M, De Smedt M, Waegemans E, Vandekerckhove B et al. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood 2011; 117: 4449–4459.
Della Gatta G, Palomero T, Perez-Garcia A, Ambesi-Impiombato A, Bansal M, Carpenter ZW et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med 2012; 18: 436–440.
Allen TD, Zhu YX, Hawley TS, Hawley RG . TALE homeoproteins as HOX11-interacting partners in T-cell leukemia. Leuk Lymphoma 2000; 39: 241–256.
Milech N, Gottardo NG, Ford J, D'Souza D, Greene WK, Kees UR et al. MEIS proteins as partners of the TLX1/HOX11 oncoprotein. Leuk Res 2010; 34: 358–363.
Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073.
Durinck K, Wallaert A, Van de Walle I, Van Loocke W, Volders PJ, Vanhauwaert S et al. The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia. Haematologica 2014; 99: 1808–1816.
Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 2014; 20: 1130–1137.
Acknowledgements
We would like to thank following funding agencies: the Fund for Scientific Research Flanders (‘FWO Vlaanderen’ research projects G.0202.09, G.0869.10N, 3G055013N, 3G056413N to FS; 3GA00113N, 3G065614, G.0C47.13N to PVV and G0B2913N, G037514N, 3G002711 to TT; doctoral grant to AW, postdoctoral grants to IvdW, TT, PVV and PR; BP is a senior clinical investigator), IWT Vlaanderen (PhD grant to KD); Ghent University (GOA grant 01G01910 to FS), the Cancer Plan from the Federal Public Service of Health (FS), the Children Cancer Fund Ghent (FS) and the Belgian Program of Interuniversity Poles of Attraction (IUAP P7/03 and P7/07). Additional funding was provided by the Cancéropole IDF, the CIT program from the Ligue Contre le Cancer, ERC St Grant Consolidator 311660, and the ANR-10-IBHU-0002 Saint-Louis Institute program (JS). We also would like to thank Aline Eggermont for excellent technical assistance.
Author contributions
KD performed and analyzed experiments and wrote the paper. WVL and MO performed bioinformatics on large-scale data sets. JVdM, IVdW, PR, AW and CEdB performed experiments. JC, JS, BP, NVR, TT, FS and PVV designed the experiments, directed research, analyzed data and wrote the paper. All the authors read and edited the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Durinck, K., Van Loocke, W., Van der Meulen, J. et al. Characterization of the genome-wide TLX1 binding profile in T-cell acute lymphoblastic leukemia. Leukemia 29, 2317–2327 (2015). https://doi.org/10.1038/leu.2015.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.162
This article is cited by
-
FAT1 expression in T-cell acute lymphoblastic leukemia (T-ALL) modulates proliferation and WNT signaling
Scientific Reports (2023)
-
The role of long non-coding RNA BCAR4 in human cancers
Human Cell (2021)
-
A novel TLX1-driven T-ALL zebrafish model: comparative genomic analysis with other leukemia models
Leukemia (2020)
-
In silico discovery of a FOXM1 driven embryonal signaling pathway in therapy resistant neuroblastoma tumors
Scientific Reports (2018)
-
Targeting BET proteins improves the therapeutic efficacy of BCL-2 inhibition in T-cell acute lymphoblastic leukemia
Leukemia (2017)