Abstract
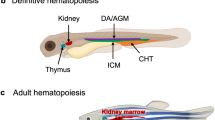
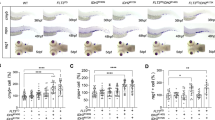
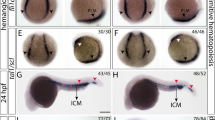
Controlled self-renewal and differentiation of hematopoietic stem/progenitor cells (HSPCs) are critical for vertebrate development and survival. These processes are tightly regulated by the transcription factors, signaling molecules and epigenetic factors. Impaired regulations of their function could result in hematological malignancies. Using a large-scale zebrafish N-ethyl-N-nitrosourea mutagenesis screening, we identified a line named LDD731, which presented significantly increased HSPCs in hematopoietic organs. Further analysis revealed that the cells of erythroid/myeloid lineages in definitive hematopoiesis were increased while the primitive hematopoiesis was not affected. The homozygous mutation was lethal with a median survival time around 14–15 days post fertilization. The causal mutation was located by positional cloning in the c-cbl gene, the human ortholog of which, c-CBL, is found frequently mutated in myeloproliferative neoplasms (MPN) or acute leukemia. Sequence analysis showed the mutation in LDD731 caused a histidine-to-tyrosine substitution of the amino acid codon 382 within the RING finger domain of c-Cbl. Moreover, the myeloproliferative phenotype in zebrafish seemed dependent on the Flt3 (fms-like tyrosine kinase 3) signaling, consistent with that observed in both mice and humans. Our study may shed new light on the pathogenesis of MPN and provide a useful in vivo vertebrate model of this syndrome for screening drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Meier B, Burton JH . Myeloproliferative disorders. Emerg Med Clin North Am 2014; 32: 597–612.
Klco JM, Vij R, Kreisel FH, Hassan A, Frater JL . Molecular pathology of myeloproliferative neoplasms. Am J Clin Pathol 2010; 133: 602–615.
Tefferi A, Vainchenker W . Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol 2011; 29: 573–582.
Langabeer SE, Haslam K, McMahon C . The molecular landscape of childhood myeloproliferative neoplasms. Leuk Res 2014; 38: 997–998.
Wilkinson AC, Gottgens B . Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol 2013; 786: 187–212.
Molofsky AV, Pardal R, Morrison SJ . Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol 2004; 16: 700–707.
Wilson A, Laurenti E, Trumpp A . Balancing dormant and self-renewing hematopoietic stem cells. Curr Opin Genet Dev 2009; 19: 461–468.
Warr MR, Pietras EM, Passegue E . Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med 2011; 3: 681–701.
de Jong JL, Zon LI . Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet 2005; 39: 481–501.
Bertrand JY, Traver D . Hematopoietic cell development in the zebrafish embryo. Curr Opin Hematol 2009; 16: 243–248.
Martin CS, Moriyama A, Zon LI . Hematopoietic stem cells, hematopoiesis and disease: lessons from the zebrafish model. Genome Med 2011; 3: 83.
Galloway JL, Zon LI . Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Curr Top Dev Biol 2003; 53: 139–158.
Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 2006; 25: 963–975.
Kissa K, Murayama E, Zapata A, Cortes A, Perret E, Machu C et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 2008; 111: 1147–1156.
Jin H, Xu J, Wen Z . Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 2007; 109: 5208–5214.
Bahary N, Davidson A, Ransom D, Shepard J, Stern H, Trede N et al. The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol 2004; 77: 305–329.
Knapik EW, Goodman A, Ekker M, Chevrette M, Delgado J, Neuhauss S et al. A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat Genet 1998; 18: 338–343.
Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S et al. Zebrafish genetic map with 2000 microsatellite markers. Genomics 1999; 58: 219–232.
Thisse C, Thisse B . High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 2008; 3: 59–69.
Yue R, Li H, Liu H, Li Y, Wei B, Gao G et al. Thrombin receptor regulates hematopoiesis and endothelial-to-hematopoietic transition. Dev Cell 2012; 22: 1092–1100.
Greig KT, Carotta S, Nutt SL . Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol 2008; 20: 247–256.
Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell 2002; 1: 433–443.
Hexner EO, Serdikoff C, Jan M, Swider CR, Robinson C, Yang S et al. Lestaurtinib (CEP701) is a JAK2 inhibitor that suppresses JAK2/STAT5 signaling and the proliferation of primary erythroid cells from patients with myeloproliferative disorders. Blood 2008; 111: 5663–5671.
Chan J, Bayliss PE, Wood JM, Roberts TM . Dissection of angiogenic signaling in zebrafish using a chemical genetic approach. Cancer Cell 2002; 1: 257–267.
Ofran Y . Concealed dagger in FLT3/ITD+ AML. Blood 2014; 124: 2317–2319.
Xie HM, Gao L, Wang N, Xu YY, Shi JL, Yu L et al. FLT3 gene overexpression and its clinical significance in acute myeloid leukemia with AML1/ETO fusion gene positive. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2014; 22: 1199–1205.
Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood 2007; 110: 1004–1012.
He BL, Shi X, Man CH, Ma AC, Ekker SC, Chow HC et al. Functions of flt3 in zebrafish hematopoiesis and its relevance to human acute myeloid leukemia. Blood 2014; 123: 2518–2529.
Oggier DM, Lenard A, Kury M, Hoeger B, Affolter M, Fent K . Effects of the protein kinase inhibitor PKC412 on gene expression and link to physiological effects in zebrafish Danio rerio eleuthero-embryos. Toxicol Sci 2011; 119: 104–115.
Nau MM, Lipkowitz S . Comparative genomic organization of the cbl genes. Gene 2003; 308: 103–113.
Rathinam C, Thien CB, Langdon WY, Gu H, Flavell RA . The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev 2008; 22: 992–997.
Thien CB, Langdon WY . Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol 2001; 2: 294–307.
Deshaies RJ, Joazeiro CA . RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78: 399–434.
Allen C, Hills RK, Lamb K, Evans C, Tinsley S, Sellar R et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013; 27: 1891–1901.
Ogawa S, Sanada M, Shih LY, Suzuki T, Otsu M, Nakauchi H et al. Gain-of-function c-CBL mutations associated with uniparental disomy of 11q in myeloid neoplasms. Cell Cycle 2010; 9: 1051–1056.
Adelaide J, Gelsi-Boyer V, Rocquain J, Carbuccia N, Birnbaum DJ, Finetti P et al. Gain of CBL-interacting protein, a possible alternative to CBL mutations in myeloid malignancies. Leukemia 2010; 24: 1539–1541.
Bandi SR, Brandts C, Rensinghoff M, Grundler R, Tickenbrock L, Kohler G et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood 2009; 114: 4197–4208.
Aranaz P, Migueliz I, Hurtado C, Erquiaga I, Larrayoz MJ, Calasanz MJ et al. CBL RING finger deletions are common in core-binding factor acute myeloid leukemias. Leuk Lymphoma 2013; 54: 428–431.
Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 2009; 460: 904–908.
Caligiuri MA, Briesewitz R, Yu J, Wang L, Wei M, Arnoczky KJ et al. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood 2007; 110: 1022–1024.
Abbas S, Rotmans G, Lowenberg B, Valk PJ . Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica 2008; 93: 1595–1597.
Reindl C, Quentmeier H, Petropoulos K, Greif PA, Benthaus T, Argiropoulos B et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res 2009; 15: 2238–2247.
Mahfouz RA, Hoteit R, Salem Z, Bazarbachi A, Mugharbel A, Farhat F et al. JAK2 V617F gene mutation in the laboratory work-up of myeloproliferative disorders: experience of a major referral center in Lebanon. Genet Test Mol Biomarkers 2011; 15: 263–265.
Kales SC, Ryan PE, Nau MM, Lipkowitz S . Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res 2010; 70: 4789–4794.
Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 2009; 113: 6182–6192.
Schnittger S, Bacher U, Alpermann T, Reiter A, Ulke M, Dicker F et al. Use of CBL exon 8 and 9 mutations in diagnosis of myeloproliferative neoplasms and myelodysplastic/myeloproliferative disorders: an analysis of 636 cases. Haematologica 2012; 97: 1890–1894.
Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet 2010; 42: 794–800.
Muramatsu H, Makishima H, Jankowska AM, Cazzolli H, O'Keefe C, Yoshida N et al. Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood 2010; 115: 1969–1975.
Shiba N, Kato M, Park MJ, Sanada M, Ito E, Fukushima K et al. CBL mutations in juvenile myelomonocytic leukemia and pediatric myelodysplastic syndrome. Leukemia 2010; 24: 1090–1092.
Hyakuna N, Muramatsu H, Higa T, Chinen Y, Wang X, Kojima S . Germline mutation of CBL is associated with moyamoya disease in a child with juvenile myelomonocytic leukemia and Noonan syndrome-like disorder. Pediatr Blood Cancer 2014; 62: 542–544.
Perez B, Mechinaud F, Galambrun C, Ben Romdhane N, Isidor B, Philip N et al. Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J Med Genet 2010; 47: 686–691.
Ogawa S, Shih LY, Suzuki T, Otsu M, Nakauchi H, Koeffler HP et al. Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res 2010; 16: 3825–3831.
Rathinam C, Thien CB, Flavell RA, Langdon WY . Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell 2010; 18: 341–352.
Dikic I, Szymkiewicz I, Soubeyran P . Cbl signaling networks in the regulation of cell function. Cell Mol Life Sci 2003; 60: 1805–1827.
Deeb KK, Smonskey MT, DeFedericis H, Deeb G, Sait SN, Wetzler M et al. Deletion and deletion/insertion mutations in the juxtamembrane domain of the FLT3 gene in adult acute myeloid leukemia. Leuk Res Rep 2014; 3: 86–89.
Beer PA, Delhommeau F, LeCouedic JP, Dawson MA, Chen E, Bareford D et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 2010; 115: 2891–2900.
Tefferi A . Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010; 24: 1128–1138.
Guglielmelli P, Vannucchi AM . Recent advances in diagnosis and treatment of chronic myeloproliferative neoplasms. F1000 Med Rep 2010; 2: pii: 16.
Konig H, Levis M . Targeting FLT3 to treat leukemia. Exp Opin Ther Targets 2014; 19: 37–54.
Annesley CE, Brown P . The biology and targeting of FLT3 in pediatric leukemia. Front Oncol 2014; 4: 263.
Acknowledgements
We thank Profs Zhu Chen for carefully revising the paper and guiding the work and Qiang Li critical reading of the manuscript, all members of the Laboratory of Development and Disease (LDD) for their technical assistance and helpful discussions, and for Drs Anskar Leung and Bai-Liang He for sharing the flt3 morpholino. This work was supported in part by Chinese National Key Basic Research Project (2013CB966800, 2011CB964803), Special Grant of Ministry of Health (201202003), Mega-projects of Science Research for the 12th Five-Year Plan (2013ZX09303302) and the National Natural Science Foundation of China (81123005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Peng, X., Dong, M., Ma, L. et al. A point mutation of zebrafish c-cbl gene in the ring finger domain produces a phenotype mimicking human myeloproliferative disease. Leukemia 29, 2355–2365 (2015). https://doi.org/10.1038/leu.2015.154
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.154