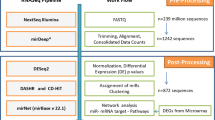
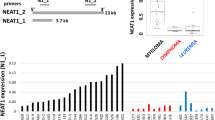
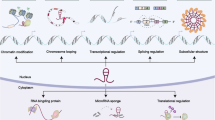
Abstract
Mutations of the tumor suppressor p53 lead to chemotherapy resistance and a dismal prognosis in chronic lymphocytic leukemia (CLL). Whereas p53 targets are used to identify patient subgroups with impaired p53 function, a comprehensive assessment of non-coding RNA targets of p53 in CLL is missing. We exploited the impaired transcriptional activity of mutant p53 to map out p53 targets in CLL by small RNA sequencing. We describe the landscape of p53-dependent microRNA/non-coding RNA induced in response to DNA damage in CLL. Besides the key p53 target miR-34a, we identify a set of p53-dependent microRNAs (miRNAs; miR-182-5p, miR-7-5p and miR-320c/d). In addition to miRNAs, the long non-coding RNAs (lncRNAs) nuclear enriched abundant transcript 1 (NEAT1) and long intergenic non-coding RNA p21 (lincRNA-p21) are induced in response to DNA damage in the presence of functional p53 but not in CLL with p53 mutation. Induction of NEAT1 and lincRNA-p21 are closely correlated to the induction of cell death after DNA damage. We used isogenic lymphoma cell line models to prove p53 dependence of NEAT1 and lincRNA-p21. The current work describes the p53-dependent miRNome and identifies lncRNAs NEAT1 and lincRNA-p21 as novel elements of the p53-dependent DNA damage response machinery in CLL and lymphoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Vogelstein B, Lane D, Levine AJ . Surfing the p53 network. Nature 2000; 408: 307–310.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Wade M, Li YC, Wahl GM . MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96.
Te Raa GD, Malcikova J, Mraz M, Trbusek M, Le Garff-Tavernier M, Merle-Beral H et al. Assessment of TP53 functionality in chronic lymphocytic leukaemia by different assays; an ERIC-wide approach. Br J Haematol 2014; 167: 565–569.
te Raa GD, Malcikova J, Pospisilova S, Trbusek M, Mraz M, Garff-Tavernier ML et al. Overview of available p53 function tests in relation to TP53 and ATM gene alterations and chemoresistance in chronic lymphocytic leukemia. Leuk Lymphoma 2013; 54: 1849–1853.
Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S . From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50.
Malcikova J, Smardova J, Rocnova L, Tichy B, Kuglik P, Vranova V et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood 2009; 114: 5307–5314.
Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 2008; 112: 3322–3329.
Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res 2009; 15: 995–1004.
Friedman RC, Farh KK, Burge CB, Bartel DP . Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105.
Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing. Cell Cycle 2007; 6: 1586–1593.
Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007; 26: 745–752.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007; 26: 731–743.
He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 2008; 582: 1564–1568.
Tazawa H, Tsuchiya N, Izumiya M, Nakagama H . Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA 2007; 104: 15472–15477.
Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007; 17: 1298–1307.
Zenz T, Mohr J, Eldering E, Kater AP, Buhler A, Kienle D et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 2009; 113: 3801–3808.
Mraz M, Malinova K, Kotaskova J, Pavlova S, Tichy B, Malcikova J et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 2009; 23: 1159–1163.
Dufour A, Palermo G, Zellmeier E, Mellert G, Duchateau-Nguyen G, Schneider S et al. Inactivation of TP53 correlates with disease progression and low miR-34a expression in previously treated chronic lymphocytic leukemia patients. Blood 2013; 121: 3650–3657.
Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD et al. Karyotype-specific microRNAsignature in chronic lymphocytic leukemia. Blood 2009; 114: 3872–3879.
Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA 2011; 305: 59–67.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–227.
Kung JT, Colognori D, Lee JT . Long noncoding RNAs: past, present, and future. Genetics 2013; 193: 651–669.
Gutschner T, Diederichs S . The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012; 9: 703–719.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142: 409–419.
Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43: 621–629.
Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 2014; 5: 5812.
Hullein J, Jethwa A, Stolz T, Blume C, Sellner L, Sill M et al. Next-generation sequencing of cancer consensus genes in lymphoma. Leuk Lymphoma 2013; 54: 1831–1835.
Castro F, Dirks WG, Fahnrich S, Hotz-Wagenblatt A, Pawlita M, Schmitt M . High-throughput SNP-based authentication of human cell lines. Int J Cancer 2013; 132: 308–314.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87.
Jethwa A, Hullein J, Stolz T, Blume C, Sellner L, Jauch A et al. Targeted resequencing for analysis of clonal composition of recurrent gene mutations in chronic lymphocytic leukaemia. Br J Haematol 2013; 163: 496–500.
Rosenkranz D, Zischler H . proTRAC—a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics 2012; 13: 5.
Ernst P, Glatting KH, Suhai S . A task framework for the web interface W2H. Bioinformatics 2003; 19: 278–282.
Anders S, Huber W . Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106.
McCarthy DJ, Chen Y, Smyth GK . Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–4297.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Jima DD, Zhang J, Jacobs C, Richards KL, Dunphy CH, Choi WW et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood 2010; 116: e118–e127.
Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K et al. MicroRNA-150 contributes to the proficiency of B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1 genes. Blood 2014; 124: 84–95.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129: 1401–1414.
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47: 648–655.
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33: 717–726.
Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood 2009; 114: 2589–2597.
Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 2010; 116: 945–952.
Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793–1801.
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol Cancer 2014; 13: 109.
Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW . Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle 2011; 10: 4237–4249.
Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 2013; 32: 5078–5088.
Wahlestedt C . Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013; 12: 433–446.
Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M et al. Characterization of the p53 cistrome—DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet 2013; 9: e1003726.
Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 2014; 3: e02200.
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006; 124: 207–219.
Wang B, Niu D, Lam TH, Xiao Z, Ren EC . Mapping the p53 transcriptome universe using p53 natural polymorphs. Cell Death Differ 2014; 21: 521–532.
Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T . Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 2012; 31: 4020–4034.
Fox AH, Lamond AI . Paraspeckles. Cold Spring Harb Perspect Biol 2010; 2: a000687.
Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y . Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 2010; 9: 1568–1576.
Rajesh C, Baker DK, Pierce AJ, Pittman DL . The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res 2011; 39: 132–145.
Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS . The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 2013; 288: 24731–24741.
Yuan M, Eberhart CG, Kai M . RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 2014; 5: 2820–2826.
Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014; 5: 5383.
Acknowledgements
This study was supported by research grants of the German Cancer Aid, GIF and the MDACC DKFZ SINF program. We thank the European Research Initiative on CLL (ERIC) for providing patient samples, HJ Delecluse and M Rogers for providing cell lines and the DKFZ Genomics and Proteomics facility for sequencing services.
Author Contributions
CJB designed and performed research, analyzed data and wrote the paper; TH and AH-W analyzed data; JH and AJ designed and performed research and analyzed data; TS, KL and SCA performed research; CCO provided IGHV mutation data; AJ provided FISH data; LS, MS, AB, AH, SD, PD, MO and OM contributed to data interpretation; DR, SD, LS, PD, TZ and AK provided patient samples; TZ designed research, interpreted data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Blume, C., Hotz-Wagenblatt, A., Hüllein, J. et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia 29, 2015–2023 (2015). https://doi.org/10.1038/leu.2015.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.119
This article is cited by
-
Expression and functions of long non-coding RNA NEAT1 and isoforms in breast cancer
British Journal of Cancer (2022)
-
Liquid–liquid phase separation drives cellular function and dysfunction in cancer
Nature Reviews Cancer (2022)
-
Targeting mutant p53 for cancer therapy: direct and indirect strategies
Journal of Hematology & Oncology (2021)
-
Circulating long non-coding RNAs HOTAIR, Linc-p21, GAS5 and XIST expression profiles in diffuse large B-cell lymphoma: association with R-CHOP responsiveness
Scientific Reports (2021)
-
Interplay between HMGA and TP53 in cell cycle control along tumor progression
Cellular and Molecular Life Sciences (2021)