Abstract
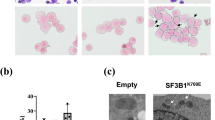
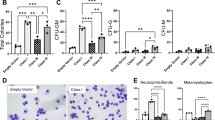
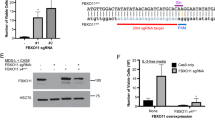
Perturbation in iron homeostasis is a hallmark of some hematologic diseases. Abnormal sideroblasts with accumulation of iron in the mitochondria are named ring sideroblasts (RS). RS is a cardinal feature of refractory anemia with RS (RARS) and RARS with marked thrombocytosis (RARS/-T). Mutations in SF3B1, a member of the RNA splicing machinery are frequent in RARS/-T and defects of this gene were linked to RS formation. Here we showcase the differences in iron architecture of SF3B1-mutant and wild-type (WT) RARS/-T and provide new mechanistic insights by which SF3B1 mutations lead to differences in iron. We found higher iron levels in SF3B1 mutant vs WT RARS/-T by transmission electron microscopy/spectroscopy/flow cytometry. SF3B1 mutations led to increased iron without changing the valence as shown by the presence of Fe2+ in mutant and WT. Reactive oxygen species and DNA damage were not increased in SF3B1-mutant patients. RNA-sequencing and Reverse transcriptase PCR showed higher expression of a specific isoform of SLC25A37 in SF3B1-mutant patients, a crucial importer of Fe2+ into the mitochondria. Our studies suggest that SF3B1 mutations contribute to cellular iron overload in RARS/-T by deregulating SLC25A37.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bjorkman SE . Chronic refractory anemia with sideroblastic bone marrow; a study of four cases. Blood 1956; 11: 250–259.
Dorion RP, Alomari M, Wood GC . Predicting the presence or absence of ringed sideroblasts in patients suspected of having a myelodysplastic syndrome and increased iron stores: a simple observation. Leukemia 2001; 15: 1793–1795.
Pellagatti A, Cazzola M, Giagounidis AA, Malcovati L, Porta MG, Killick S et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood 2006; 108: 337–345.
Boultwood J, Pellagatti A, Nikpour M, Pushkaran B, Fidler C, Cattan H et al. The role of the iron transporter ABCB7 in refractory anemia with ring sideroblasts. PLoS One 2008; 3: e1970.
Cazzola M, Invernizzi R . Ring sideroblasts and sideroblastic anemias. Haematologica 2011; 96: 789–792.
Sato K, Torimoto Y, Hosoki T, Ikuta K, Takahashi H, Yamamoto M et al. Loss of ABCB7 gene: pathogenesis of mitochondrial iron accumulation in erythroblasts in refractory anemia with ringed sideroblast with isodicentric (X)(q13). Int J Hematol 2011; 93: 311–318.
Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L et al. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood 2007; 110: 1353–1358.
Caudill JS, Imran H, Porcher JC, Steensma DP . Congenital sideroblastic anemia associated with germline polymorphisms reducing expression of FECH. Haematologica 2008; 93: 1582–1584.
Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 2012; 26: 542–545.
Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011; 365: 1384–1395.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69.
Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood 2012; 120: 3173–3186.
Williams D, Carter CB . Transmission Electron Microscopy: A Textbook For Material Science. Chapter 39 (High-Energy Loss Spectra and Images). Plenum Press: New York, USA, 1996; pp 715–716.
Tan H, Verbeeck J, Abakumov A, Van Tendeloo G . Oxidation state and chemical shift investigation in transition metal oxides by EELS. Ultramicroscopy 2012; 116: 24–33.
Hirayama T, Okuda K, Nagasawa H . A highly selective turn-on fluorescent probe for iron(II) to visualize labile iron in living cells. Chem Sci 2013; 4: 1250–1256.
Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia 2013; 28: 78–87.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B . Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628.
Oshlack A, Robinson MD, Young MD . From RNA-seq reads to differential expression results. Genome Biol 2010; 11: 220.
Smyth G . Limma: linear models for microarray data. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds.) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer: New York, USA, 2005; pp 397–420.
Young MD, Wakefield MJ, Smyth GK, Oshlack A . Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11: R14.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21.
Glaus P, Honkela A, Rattray M . Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics 2012; 28: 1721–1728.
Barnard J, Rubin DB . Miscellanea. Small-sample degrees of freedom with multiple imputation. Biometrika 1999; 86: 948–955.
Wang Y, Langer NB, Shaw GC, Yang G, Li L, Kaplan J et al. Abnormal mitoferrin-1 expression in patients with erythropoietic protoporphyria. Exp Hematol 2011; 39: 784–794.
Garvie LAJ, Buseck PR . Ratios of ferrous to ferric iron from nanometre-sized areas in minerals. Nature 1998; 396: 667–670.
Visconte V, Mahfouz RZ, Tabarroki A, Hasrouni E, Rogers HJ, Saunthararajah Y et al. BCL-2 family of genes is a key regulator in the pathogenesis of SF3B1 mutant and wild type MDS with ring sideroblasts and represents a novel drug target in this disease. (ASH Annual Meeting). Blood 2013; 122: 263.
Chen W, Dailey HA, Paw BH . Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 2010; 116: 628–630.
Visconte V, Mahfouz RZ, Barnard J, Tabarroki A, Zhang L, Hasrouni E et al. Splicing factor 3b subunit 1 (SF3B1) mediates mitochondrial iron overload in myelodysplastic syndromes with ring sideroblasts by alternative splicing of mitoferrin-1 (SLC25A37). (ASH Annual Meeting). Blood 2013; 122: 1555.
Visconte V, Makishima H, Maciejewski JP, Tiu RV . Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia 2012; 26: 2447–2454.
Visconte V, Tabarroki A, Rogers HJ, Hasrouni E, Traina F, Makishima H et al. SF3B1 mutations are infrequently found in non-myelodysplastic bone marrow failure syndromes and mast cell diseases but, if present, are associated with the ring sideroblast phenotype. Haematologica 2013; 98: e105–e107.
Nikpour M, Scharenberg C, Liu A, Conte S, Karimi M, Mortera-Blanco T et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 2013; 27: 889–896.
Gattermann N . SF3B1 and the riddle of the ring sideroblast. Blood 2012; 120: 3167–3168.
Rademakers LH, Koningsberger JC, Sorber CW, Baart de la Faille H, Van Hattum J, Marx JJ . Accumulation of iron in erythroblasts of patients with erythropoietic protoporphyria. Eur J Clin Invest 1993; 23: 130–138.
Grasso JA, Myers TJ, Hines JD, Sullivan AL . Energy-dispersive X-ray analysis of the mitochondria of sideroblastic anaemia. Br J Haematol 1980; 46: 57–72.
Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J . Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol 2009; 29: 1007–1016.
Michael IP, Kurlender L, Memari N, Yousef GM, Du D, Grass L et al. Intron retention: a common splicing event within the human kallikrein gene family. Clin Chem 2005; 51: 506–515.
Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ . Detection and evaluation of intron retention events in the human transcriptome. RNA 2004; 10: 757–765.
Chen W, Paradkar PN, Li L, Pierce EL, Langer NB, Takahashi-Makise N et al. Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci USA 2009; 106: 16263–16268.
Lane DJ, Richardson DR . Frataxin, a molecule of mystery: trading stability for function in its iron-binding site. Biochem J 2010; 426: e1–e3.
Acknowledgements
We would like to thank Dr Andrea N. Ladd of the Department of Cellular and Molecular Medicine, Cleveland Clinic Lerner Institute, Cleveland OH and Dr Velizar Shivarov of the Laboratory of Hematopathology and Immunology, National Hematology Hospital, Sofia, Bulgaria for their helpful recommendations and careful revisions of the manuscript. This work was supported in full or partially by Cleveland Clinic Seed Support, and Scott Hamilton CARES grant (RVT)
Author contributions
VV conceived the study, performed experiments, analyzed the data and wrote the manuscript; NA performed and analyzed electron microscopy experiments; RM performed experiments and provided important insights to the manuscript; AT collected data and performed experiments; JC analyzed and interpreted the electron microscopy results; JP synthesized the RhoNox-1 compound; RS-M analyzed the electron microscopy results; MH designed protocols of slides preparation for electron microscopy; HJR reviewed BM pathology and edited the manuscript; EH performed experiments; MAS provided patients; AHH provided fundamental feedbacks and long track experience on material science to the manuscript; YS provided patients and edited the manuscript; JB analyzed the RNA-Seq data and provided important suggestions; RVT conceived the study, designed the experimental approach, reviewed the clinical data, interpreted the data, provided the patients and wrote the manuscript; all authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Visconte, V., Avishai, N., Mahfouz, R. et al. Distinct iron architecture in SF3B1-mutant myelodysplastic syndrome patients is linked to an SLC25A37 splice variant with a retained intron. Leukemia 29, 188–195 (2015). https://doi.org/10.1038/leu.2014.170
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.170
This article is cited by
-
Iron overload alters the energy metabolism in patients with myelodysplastic syndromes: results from the multicenter FISM BIOFER study
Scientific Reports (2020)
-
Myelodysplastic Syndrome/Myeloproliferative Neoplasm (MDS/MPN) Overlap Syndromes: Molecular Pathogenetic Mechanisms and Their Implications
Indian Journal of Hematology and Blood Transfusion (2019)
-
Turning the tide in myelodysplastic/myeloproliferative neoplasms
Nature Reviews Cancer (2017)
-
Splicing factor mutations in MDS RARS and MDS/MPN-RS-T
International Journal of Hematology (2017)
-
Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes
Leukemia (2016)