Abstract
The Notch signaling pathway has been recognized as a key factor for the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL), because of the high incidence of activating mutations of Notch1. Notch inhibition could serve as a new treatment strategy for T-ALL; however, the attempts to perturb Notch signaling pathways have been unsuccessful so far. In this study, we found that proteasome inhibitors exert cytotoxic effects on T-ALL cells with constitutive activation of Notch1 to a similar extent as myeloma cells. The proteasome inhibitor bortezomib repressed the transcription of Notch1 and downstream effectors including Hes1, GATA3, RUNX3 and nuclear factor-κB (NF-κB) (p65 and p50), coincided with downregulation of the major transactivator Sp1 and its dissociation from Notch1 promoter. Overexpression of the Notch1 intracellular domain (NICD) significantly ameliorated bortezomib-induced cytotoxicity against T-ALL cells. Drug combination studies revealed that bortezomib showed synergistic or additive effects with key drugs for the treatment of T-ALL such as dexamethasone (DEX), doxorubicin and cyclophosphamide, which were readily abolished by NICD overexpression. The synergy of bortezomib and DEX was confirmed in vivo using a murine xenograft model. Our findings provide a molecular basis and rationale for the inclusion of proteasome inhibitors in treatment strategies for T-ALL.
Similar content being viewed by others
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic malignancy of thymocytes. The disease represents approximately 15% of pediatric and 25% of adult ALL cases. Despite the application of intensive multiagent chemotherapy, the treatment outcome of T-ALL is not satisfactory and the prognosis of relapsed patients is still very poor.1 Given the limitation of current approaches, it is important to develop new treatment strategies based on increased understanding of the pathophysiology of T-ALL.
Notch1 has been recognized as one of the most important genes in T-ALL pathogenesis, because activating mutations of Notch1 are detected in >50% of T-ALL patients2, 3, 4 and Notch1 overexpression transforms normal hematopoietic stem cells and T-cell progenitors to T-ALL in murine models.5 Recent investigations suggest that Notch1 is also implicated in drug sensitivity of T-ALL cells.6, 7 It has been reported that the pre-treatment response to glucocorticoids is a major independent factor influencing the survival of ALL patients8 and the sensitivity to glucocorticoids is negatively correlated with the expression levels of Notch1 in T-ALL cells.9
Notch1 is one of four single transmembrane Notch receptors composed of extracellular and intracellular domains that are non-covalently linked via the heterodimerization domains.10, 11 In response to the binding of cognate ligands, Notch receptors are subjected to proteolytic cleavage by γ-secretase, resulting in the liberation of the Notch1 intracellular domain (NICD). Subsequently, NICD translocates to the nuclei and activates the transcription of various genes involved in cell cycle progression, cellular differentiation and apoptosis inhibition.11, 12 In T-ALL, the mutations of Notch1 occur in the proline/glutamic acid/serine/threonine-rich and/or heterodimerization domains to cause constitutive activation of Notch1 via stabilization of NICD and/or ligand-independent generation of NICD.2, 11, 12
On the basis of these findings, γ-secretase inhibitors (GSIs) were expected to be effective for T-ALL because of their ability to block the generation of NICD.13, 14, 15 Unfortunately, most T-ALL cells, especially those with heterodimerization domain mutations, are resistant to GSIs.2, 16 Indeed, clinical trials disclosed the limitation of antitumor activity and a number of severe adverse effects of GSIs.17, 18 In addition, blocking antibodies for specific Notch receptors and/or ligands are also found to be of limited value for the treatment of T-ALL associated with aberrant Notch1 activation.19, 20, 21 Therefore, the development of novel agents targeting the Notch-mediated oncogenic pathway is eagerly awaited.
Proteasome inhibition is now considered a unique and effective way to kill cancer cells that can tolerate conventional chemotherapy. Bortezomib is the first proteasome inhibitor approved for clinical application and is now widely used for the treatment of multiple myeloma (MM).22 Previous studies showed that bortezomib affects several molecules including nuclear factor-κB (NF-κB) and histone deacetylases (HDACs) in MM cells,23, 24 both of which could be therapeutic targets of T-ALL.25, 26, 27, 28 However, relatively little is known about the cytotoxic activity of bortezomib against T-ALL and its mechanisms of action, although a preliminary report suggests the possibility.29 In this study, we demonstrate that proteasome inhibitors, bortezomib and K-7174, exert marked cytotoxicity against T-ALL cells in vitro and in vivo, and also increase the sensitivity of T-ALL cells to conventional chemotherapeutic agents via transcriptional repression of Notch1 and downstream effectors such as NF-κB and RUNX3. These findings provide a rationale for the inclusion of proteasome inhibitors in treatment strategies for T-ALL.
Materials and methods
Cells and cell culture
We used four bona fide human T-ALL cell lines, Jurkat, CEM, MOLT4 and KOPT-K1 (provided by Dr Takeshi Inukai, University of Yamanashi, Yamanashi, Japan), in this study.2 Other cell lines and their origins are KMS12-BM, U266, RPMI8226 (MM), KOPM30 (B-ALL), HBL-2 (mantle cell lymphoma), Namalwa (Burkitt lymphoma), HL-60 and K562 (acute myeloid leukemia), all of which were purchased from the Health Science Research Resources Bank (Osaka, Japan).
Drugs
The drugs used in this study and their sources are bortezomib, MLN120B (Millennium Pharmaceuticals, Cambridge, MA, USA), K-7174 (Kowa, Tokyo, Japan), vincristine (Shionogi, Osaka, Japan), doxorubicin (ADM) (Meiji, Tokyo, Japan), mithramycin, dexamethasone (DEX) (Sigma-Aldrich, St Louis, MO, USA), cytosine arabinoside and 4-hydroxycyclophospamide (Wako Biochemicals, Osaka, Japan). All drugs were dissolved in dimethyl sulfoxide at appropriate concentrations and used at a final dilution of 1/1000.
Cell proliferation assays
Cell proliferation was monitored using a Cell Counting Kit (Wako Biochemicals). In brief, cells were seeded in 96-well flat-bottomed microplates at a density of 1 × 105 per well and incubated with or without drugs at 37 °C. After incubation, the absorbance was measured at a wavelength of 450 nm using a microplate reader, and expressed as a percentage of the value of corresponding untreated cells.24
Assessment of cell death
Cells were washed with phosphate-buffered saline and stained with phycoerythrin-conjugated annexin-V (annexin-V/PE) (Biovision, Mountain View, CA, USA). Cell death/apoptosis was judged by annexin-V reactivity using a BD LSRFortessa flow cytometer (Becton Dickinson, Bedford, MA, USA).24
Drug combination study
We calculated the combination index of bortezomib and other anti-leukemic drugs using the CompuSyn software and generated isobolograms according to the manufacturer’s instructions (www.combosyn.com). The overall effects of drug combination were analyzed by the method of Chou and Talalay.30
Real-time quantitative reverse transcriptase-PCR
Total cellular RNA was isolated from 1 × 105 cells using an RNeasy Kit (Qiagen, Valencia, CA, USA) and reverse-transcribed into complementary DNA using ReverTra Ace and oligo (dT) primers (Toyobo, Tokyo, Japan). We performed real-time quantitative reverse transcriptase-PCR using the Expression Assays (Hs01062014 for Notch1, Hs00172878 for HES1, Hs00211000 for CYLD, Hs00231122 for GATA3, Hs00231709 for RUNX3, Hs00153294 for RELA, Hs00765730 for NFKB1 and Hs01922876 for glyceraldehyde 3-phosphate dehydrogenase (GAPDH)) and TaqMan Fast Universal PCR Master Mix as described previously.31
Immunoblotting
Immunoblotting was carried out according to the standard method using the following antibodies: anti-Notch1, anti-cleaved Notch1, anti-KLF4, anti-p105/p50, anti-p100/p52, anti-p65, anti-c-Rel, anti-IKKβ, anti-phosphorylated IKKα/β, anti-IκBα (Cell Signaling Technology, Beverly, MA, USA), anti-HDAC1 (Sigma-Aldrich), anti-Sp1, anti-histone H1, anti-MZF-1 and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA). We used a nuclear extraction kit (Cayman Chemical, Ann Arbor, MI, USA) to separate cytoplasm and nuclear fractions.
NF-κB assay
NF-κB activity was quantitatively measured as p65 and p50 bound to κB consensus oligonucleotides (5′-AGTTGAGGGGACTTTCCCAGGC-3′) in enzyme-linked immunosorbent assay using the NF-κB Transcription Factor Assay kit (Cayman Chemical).32
Chromatin immunoprecipitation assays
We used the ChIP-IT Express Enzymatic (Active Motif, Carlsbad, CA, USA) to perform chromatin immunoprecipitation assays. In brief, we fixed cells in 1% formaldehyde at room temperature for 10 min and isolated chromatin fractions using enzymatic shearing. After centrifugation, supernatants were incubated with antibodies of interest and protein G magnetic beads at 4 °C overnight. We purified DNA fragments from the mixture according to the manufacturer’s instructions and carried out PCR using Mighty Amp (Takara, Shiga, Japan) and the primers depicted in Supplementary Table 1.
Reporter assays
We amplified the promoter regions of the Notch1 gene (–392 to –1, –342 to –1, –315 to –1 and –300 to –1) using PCR (for primers, see Supplementary Table 1) and inserted them into the pGL4.17 firefly luciferase vector (Promega, Madison, WI, USA) to generate reporter plasmids. We introduced reporter plasmids into CEM cells along with the pGL4.73 Renilla luciferase vector (Promega), which served as a positive control to determine transfection efficiencies, using electroporation. After 48 h, firefly and Renilla luciferase activities were measured discriminately using the Dual-Luciferase Reporter Assay System (Promega). The promoterless pGL4.17-basic vector was used as a negative control. Luciferase activity was normalized with the internal standard and indicated as a relative ratio to negative controls.
Lentiviral transduction of NICD in T-ALL cells
The NICD fragment of the Notch1 gene was amplified by PCR using specific primers (forward: 5′-TTCTCGAGCACCATGGTGCTGCTGTCCCGCAAGCG-3′, reverse: 5′-AAGGATCCTTACTTGAAGGCCTCCGGAATG-3′) using complementary DNA from normal T-lymphocyte,16 and inserted into the CSII-CMV-MCS-IRES2-VENUS vector (provided by Dr Hiroyuki Miyoshi, RIKEN BioResorce Center, Ibaraki, Japan). This vector was co-transfected into 293FT cells with packaging plasmids (Invitrogen, Carlsbad, CA, USA) to produce infective lentiviruses in culture supernatants. Lentiviruses were then added to cell suspensions and transduced for 24 h as previously described.24 We established stable transformants by isolating single-cell clones using limiting dilution (1 cell per well in 96-well culture plates) after long-term culture. NICD overexpression was verified by immunoblotting for each clone.
Murine xenograft experiments
We established luciferase-expressing MOLT4 sublines by transfecting the CSII-CMV-MCS-IRES2-VENUS vector-carrying firefly luciferase complementary DNA (provided by Dr Takashi Murakami, Takasaki University of Health and Welfare, Gunma, Japan). Cell suspensions (1 × 107 cells in 1 × 10−4 l of Iscove’s modified Dulbecco’s medium) were inoculated subcutaneously into the right thigh of nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Charles River Laboratories, Wilmington, MA, USA) together with 1 × 10−4 l Matrigel basement membrane matrix (Becton Dickinson).16, 24 When tumors were measurable after 3–5 days, mice were assigned to four treatment groups receiving the vehicle alone (0.9% NaCl, five times a week), bortezomib alone (0.25 mg/kg twice a week), DEX alone (10 mg/kg five times a week) or bortezomib plus DEX intraperitoneally (n=3–4 in each group). Caliper measurements of the longest perpendicular tumor diameters were performed to estimate the tumor volume using the following formula: 4/3π × (width/2)2 × (length/2), which represents the three-dimensional volume of an ellipse.
Tumor progression was also determined by measuring tumor-derived luciferase activity with the noninvasive bioimaging system. Tumor-bearing mice were injected with the luciferase substrate D-luciferin (1.5 mg/body) intraperitoneally after being anesthetized with isoflurane. Photons transmitted through the body were collected for a specified length of time and analyzed using the IVIS Imaging System with Living Image software (Xenogen, Alameda, CA, USA). Quantitative data were expressed as photon units (photons/s).
Results
Proteasome inhibition causes marked cytotoxicity in T-ALL cells
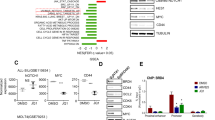
The efficacy of bortezomib for hematological malignancies other than MM and mantle cell lymphoma remains to be determined; therefore, we screened for the cytotoxic effects of bortezomib against different types of hematological malignancies. As shown in Figure 1a, bortezomib was particularly effective for T-ALL cell lines with IC50 values of 13.0±2.9, 4.5±0.24, 3.5±0.24 and 4.7±0.29 nM for Jurkat, MOLT4, CEM and KOPT-K1, respectively. The IC50 values for bortezomib were lower in T-ALL than MM cell lines except for Jurkat. Flow cytometric analysis revealed that growth inhibition was mainly due to the induction of apoptosis but not cell cycle arrest (data not shown). In addition, we found that K-7174, a homopiperazine derivative with a distinct mode of proteasome inhibition from bortezomib,33, 34 was also very effective for T-ALL cell lines including Jurkat (Figure 1a). We confirmed that bortezomib readily inhibited proteasome β5 activity in T-ALL cells in a dose-dependent manner (Supplementary Figure 1). These results suggest that proteasome inhibition has a therapeutic impact on T-ALL as well as MM.
Proteasome inhibitors are effective for T-ALL cell lines with constitutive activation of Notch1. (a) The IC50 values of bortezomib (open column) and K-7174 (filled column) were calculated from dose-response curves of cell lines from various hematological malignancies including MM (U266, KMS12-BM and RPMI 8226), B-lymphoid malignancies (KOPM30, HBL-2 and Namalwa), T-ALL (Jurkat, MOLT4, CEM and KOPT-K1) and myeloid leukemias (HL-60 and K562) obtained at 72 h of culture. (b) Whole-cell lysates were prepared from the indicated cell lines and subjected to immunoblotting using specific antibodies that recognize full-length Notch1 (FL), TM and intracellular domain of Notch1 (NICD). The membranes were reprobed with anti-GAPDH antibody to serve as a loading control.
Bortezomib downregulates the expression of Notch1 in T-ALL cells
Next, we investigated the molecular mechanisms by which proteasome inhibitors affect T-ALL cells using bortezomib, because this drug has already been approved for clinical use in MM and mantle cell lymphoma.22 The high sensitivity to bortezomib indicates that its targets are critical for T-ALL cell survival, such as Notch1,2, 16 NF-κB25, 26 and HDAC1.27, 28 During the activation of Notch signaling pathways, Notch is proteolytically processed and assembled as heterodimeric proteins composed of an extracellular ligand-binding domain, a single-pass transmembrane domain (TM) and NICD.10 As NICD in turn translocates to the nucleus and activates the transcription of downstream target genes, the amounts of nuclear NICD reflect the overall activity of Notch signaling pathways. To estimate Notch1 activity, we performed immunoblot analysis for the expression of NICD in various hematopoietic cell lines. As shown in Figure 1b, constitutive activation of Notch1 was exclusively observed in T-ALL cell lines.
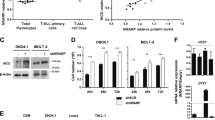
We then asked whether bortezomib could target constitutively active Notch1 in T-ALL cells. As shown in Figure 2a, bortezomib markedly inhibited Notch1 activity as evidenced by the disappearance of NICD. The downregulation of NICD was most striking with bortezomib, followed by DEX, among key anti-ALL agents at equitoxic concentrations, suggesting that bortezomib is suitable for therapeutic targeting of Notch1 in T-ALL. Bortezomib readily decreased the expression levels of not only NICD but also full-length Notch1 and TM in a time- and dose-dependent manner (Figures 2b and c). In contrast, bortezomib did not affect the expression of HDAC1, another highly expressed possible therapeutic target of T-ALL (Figure 2c). In addition, bortezomib did not upregulate the expression of CCAAT/enhancer-binding protein homologous protein (data not shown), suggesting that, unlike myeloma cells,23, 34 endoplasmic reticulum stress is not a principal mechanism of bortezomib-mediated cytotoxicity in T-ALL cells.
Bortezomib downregulates the expression of Notch1 and affects Notch signaling pathways at transcriptional levels in T-ALL cells. (a) MOLT4 cells were cultured in the absence (Control) or presence of IC50 doses of various anti-leukemia agents (1 μM 4-hydroxycyclophospamide (4-OHCY), 15 nM ADM, 150 nM DEX, 1 nM vincristine (VCR), 5 nM cytosine arabinoside (Ara-C) and 4 nM bortezomib) and subjected to immunoblotting for full-length Notch1 (FL), TM and cleaved Notch1 (NICD) after 24 h. The membranes were reprobed with anti-GAPDH antibody to serve as a loading control. (b) T-ALL cells were cultured with bortezomib at IC50 (12 nM for Jurkat, 4 nM for MOLT4 and CEM) for up to 24 h. Whole-cell lysates were prepared at the indicated time points and subjected to immunoblotting for Notch1 (FL and TM), NICD, Sp1 and GAPDH. (c) T-ALL cells were cultured with the indicated concentrations of bortezomib for 24 h, and subjected to immunoblotting for Notch1 (FL and TM), NICD, HDAC1 and GAPDH. (d) Total cellular RNA was isolated simultaneously in the experiments described in (c), and subjected to real-time quantitative reverse transcriptase (RT)-PCR. The mRNA expression of Notch1 was normalized to that of GAPDH and quantified by the 2−ΔΔCt method with the values of untreated cells being set at 1.0. The means±s.d. (bars) of three independent experiments are shown. P-values were calculated by one-way analysis of variance with Tukey’s multiple comparison test. Asterisks indicate P<0.05. (e) The mRNA expression of HES1, CYLD, GATA3 and RUNX3 was quantified by real-time quantitative RT-PCR in the indicated cell lines treated with bortezomib at IC50 (12 nM for Jurkat, 4 nM for MOLT4 and CEM) for 24 h. The means±s.d. (bars) of three independent experiments are shown. P-values were calculated by Student’s t-test. Asterisks indicate P<0.05.
Bortezomib suppresses the expression of Notch1 and its targets at transcriptional levels
Next, we determined whether the downregulation of Notch1 expression occurs at transcriptional or post-transcriptional levels in T-ALL cells. We assumed that bortezomib repressed the transcription of the Notch1 gene because of the decrease in the abundance of full-length Notch1 and TM in previous experiments. To validate this assumption, we performed real-time quantitative PCR analysis and confirmed the downregulation of Notch1 mRNA in T-ALL cells during bortezomib treatment (Figure 2d). The extent and kinetics of Notch1 mRNA decrease were well correlated with those at protein levels. We then examined the expression of Notch target genes including HES1,25, 35 GATA336 and RUNX3, which was reported to confer drug resistance to T-ALL cells via repression of RUNX1 and PKC-θ.37 As expected, the expression of Notch direct targets was downregulated by bortezomib coincided with a decrease in Notch1 expression in T-ALL cell lines, except for HES1 in CEM for unknown reasons (Figure 2e). These results imply that bortezomib mainly downregulates Notch1 and its targets at transcriptional levels and thus, would be effective for overcoming drug resistance in T-ALL cells.
Bortezomib impedes NF-κB activity by affecting the expression of p65 and p50 in T-ALL
It has been reported that NF-κB is activated via NICD/Hes1-mediated repression of the deubiquitinase CYLD and has a crucial role in the survival and drug resistance of T-ALL cells.25, 26 We therefore investigated whether bortezomib impedes NF-κB activity as a consequence of NICD downregulation. Toward this end, we first screened for the expression and localization of the components of NF-κB pathways in T-ALL cell lines. As shown in Figures 3a, p65 and p50 were detected in nuclear fractions and thus, the p65/p50 complex might be constitutively activated in Jurkat and CEM cells, whereas p65/p65 homodimer seemed to be a dominant complex in MOLT4 cells because of faint expression of p105/p50, which is consistent with previous reports.29, 38 In contrast, these cell lines did not express non-canonical NF-κB components such as c-Rel, p100 and p52.
Bortezomib inhibits canonical NF-κB activity by downregulating the expression of p65 and p50 in T-ALL cells. (a) We extracted nuclear and cytoplasmic fractions from T-ALL cells, and subjected them to immunoblotting for the components of NF-κB pathways. The quality of separation was monitored using histone H1 and GAPDH as nuclear and cytoplasmic markers, respectively. (b) We cultured T-ALL cells with bortezomib at IC50 (12 nM for Jurkat, 4 nM for MOLT4 and CEM) for the indicated periods and examined the expression of the components of NF-κB pathways using immunoblotting. (c) We cultured T-ALL cells with the indicated concentrations of bortezomib for 24 h and examined the expression of the components of NF-κB pathways using immunoblotting. The data shown are representative of multiple independent experiments. (d) We cultured T-ALL cells in the absence (open column) or presence (closed column) of bortezomib at IC50 (12 nM for Jurkat, 4 nM for MOLT4 and CEM) and prepared nuclear fractions after 24 h. The amounts of p65 and p50 bound to κB consensus sequence (5′-AGTTGAGGGGACTTTCCCAGGC-3′) was quantitatively measured by an enzyme-linked immunosorbent assay using specific antibodies against human p65 and p50.
Having determined NF-κB components expressed in T-ALL cells, we investigated the effects of bortezomib on their abundance and activity. Unexpectedly, bortezomib failed to increase the abundance of IκBα protein (Figures 3b and c), although it restored the expression of CYLD in parallel with HES1 downregulation (Figure 2e). In addition, bortezomib did not only activate IκB kinase β (IKKβ) but decreased its expression levels. In support of this observation, the IKKβ inhibitor MLN120B was rather antagonistic with bortezomib in T-ALL cells, but became synergistic when NICD was overexpressed (Supplementary Figure 2). This could be explained by the negative effect of bortezomib on IKKβ and its restoration by NICD overexpression (data not shown). Instead, bortezomib markedly reduced the amounts of p65 and p50 in a time- and dose-dependent manner (Figures 3b and c). The repression of p50 expression appeared to be mainly transcriptional, because bortezomib decreased the expressions of NFKB1 mRNA and its translational product p105 (Supplementary Figure 3 and Figures 3b and c). In contrast, p65 mRNA expression was unaffected or even increased in MOLT4 cells, implying that bortezomib perturbs the stability of p65 via NICD downregulation as suggested previously.29, 39 As a result of decreased expressions of p65 and p50, canonical NF-κB activity, as detected by the binding of nuclear p65 and p50 to the κB-binding consensus, was considerably reduced in bortezomib-treated T-ALL cells (Figure 3d).
Degradation of Sp1 protein underlies transcriptional repression of Notch1 by bortezomib
Next, we investigated the mechanisms of transcriptional repression of the Notch1 gene during bortezomib treatment in T-ALL cells. First, we performed reporter assays to determine bortezomib-responsive regions in the Notch1 promoter. Previously, Lambertini et al.40 reported that the segment between –392 and –1 of the Notch1 gene confers full promoter activity in human keratinocytes. Based on their finding, we constructed four reporter plasmids carrying a full promoter and its deletion fragments (Figure 4a, left panel) and investigated the effects of bortezomib on their activities in T-ALL cells. As shown in Figure 4a, bortezomib significantly reduced the activity of the region –392 to –1 but not others, suggesting that the bortezomib-responsive element is localized in the segment between –392 and –342. This segment is GC-rich (5′-CGGGGAGGCGCAAAGGCGGACGGGGCGTGCGGGAGGAGGTGGCCGCGGAGG-3′) and contains putative binding sites for Sp1, KLF4 and MZF-1 according to database search. Immunoblot analysis revealed that bortezomib markedly downregulated the expression of Sp1, whereas the other two transcription factors were not expressed in T-ALL cells (Figures 2b and 4b). In a previous study, we have shown that Sp1 protein is cleaved and degraded by activated caspase-8 and caspase-3 during bortezomib treatment in MM cells.24, 34 We confirmed the activation of caspase-3 in bortezomib-treated T-ALL cells (data not shown), which might be implicated in Sp1 cleavage and subsequent degradation, and partial restoration of Notch1 downregulation by a specific inhibitor of caspase-3 (Supplementary Figure 4). We further corroborated the importance of Sp1 for Notch1 transcription using mithramycin, which disturbs Sp1 binding to GC boxes and induces auto-repression of Sp1 expression.41, 42 Treatment with mithramycin strikingly reduced the abundance of full-length Notch1 and its downstream target p65 in parallel with Sp1 downregulation in T-ALL cells (Figure 4c).
Bortezomib decreases Notch1 promoter activity via Sp1 downregulation. (a) We transfected 5 μg of pGL4.17 plasmid containing Notch1 promoter sequences between –392 and –1, –342 and –1, –315 and –1, and –300 and –1, respectively, into CEM cells and measured luciferase activities in the absence (open column) or presence (closed column) of 4 nM bortezomib after 48 h. Notch1 promoter activity was calculated as relative firefly luciferase activities against untreated cells after normalization of transfection efficiencies using co-transfected Renilla luciferase activities. The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test. (b) T-ALL cells were cultured in the absence (−) or presence (+) of bortezomib at IC50 (12 nM for Jurkat, 4 nM for MOLT4 and CEM) for 24 h, and subjected to immunoblotting for Sp1, KLF4, MZF-1 and GAPDH (loading control). The antibodies against KLF4 and MZF-1 were validated using HEK293 cells transfected with a KLF4 expression vector and K562 cells as positive controls, respectively. (c) Jurkat cells were cultured with mithramycin at the indicated concentrations for 24 h and subjected to immunoblotting using specific antibodies against Notch1 (FL and TM), Sp1, p65 and GAPDH (loading control). (d) MOLT4 and CEM cells were cultured in the absence (−) or presence (+) of 4 nM bortezomib, and subjected to chromatin immunoprecipitation assays after 48 h. Chromatin suspensions were immunoprecipitated with anti-Sp1 and corresponding control antibodies (IgG). The precipitants were subjected to PCR to amplify the promoter region (–480 to –342) that includes Sp1-binding sites of the Notch1 gene. The amplified products were visualized by ethidium bromide staining after 2% agarose gel electrophoresis. Representative data of 40 cycles are shown. Input indicates that PCR was performed with genomic DNA.
Next, we performed chromatin immunoprecipitation assays to verify the binding of Sp1 to the region between –392 and –342 of Notch1 promoter. Although there are several Sp1 consensus sites in Notch1 promoter, actual binding was detected only in the region between –480 and –342 (Figure 4d and data not shown). Sp1 is dissociated from Notch1 promoter upon bortezomib treatment in MOLT4 and CEM cells, consistent with the findings of reporter assays. Taken together, Sp1 confers the baseline expression of the Notch1 gene and its downregulation by bortezomib leads to transcriptional repression of Notch1 in T-ALL cells.
Notch1 overexpression rescues T-ALL cells from bortezomib-induced apoptosis
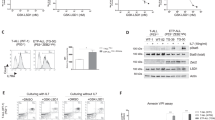
To confirm the dependency of bortezomib action on Notch1 in T-ALL cells, we performed gain-of-function analyses using Jurkat and MOLT4 cells lentivirally transduced with either mock or NICD expression vector (Figure 5a). As shown in Figure 5b, NICD overexpression almost completely abrogated bortezomib-induced apoptosis in both Jurkat and MOLT4 cells. Furthermore, we established NICD-overexpressing stable transformants and confirmed that these cells were resistant to bortezomib in terms of Notch1 downregulation (Figure 5c), NF-kB inactivation (Figures 5c and d) and cytotoxicity (Figure 5e). In addition, NICD overexpression almost completely reversed mithramycin-induced suppression of NF-κB activity (Figure 5d) and apoptosis (Figure 5e), confirming that NICD mostly acts downstream of Sp1. Moreover, NICD overexpression conferred the resistance to not only bortezomib but also other anticancer drugs such as DEX and cytosine arabinoside (Figure 5f). These results indicate that bortezomib induces marked cytotoxicity in T-ALL cells by targeting Notch1-mediated oncogenic pathways involving NF-κB and might modulates the sensitivity to other anticancer agents.
Effects of NICD overexpression on bortezomib-induced apoptosis in T-ALL cells. (a) Jurkat and MOLT4 cells were lentivirally transduced with either CSII-VENUS (mock) or CSII-VENUS-NICD vector (NICD). Whole-cell lysates were prepared from VENUS-positive cells and subjected to immunoblotting for NICD and GAPDH. (b) Mock- and NICD-transduced cells were cultured in the absence (−) or presence (+) of bortezomib at IC50 (12 nM for Jurkat and 4 nM for MOLT4) for 48 h and stained with annexin-V/PE in preparation for flow cytometric analysis. The y-axis shows the proportion of annexin-V-positive cells (%) in the VENUS-positive fraction. The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test. (c) We established stable transformants from MOLT4 cells lentivirally transduced with either mock or NICD, and examined the expression of NICD, p65 and GAPDH after 24 h of culture with the indicated concentrations of bortezomib. (d) NF-κB activity was measured by an enzyme-linked immunsorbent assay for p65 using nuclear extracts from mock- and NICD-transfected MOLT4 cells cultured in the absence (control) or presence of either 4 nM bortezomib (filled column) or 40 nM mithramycin (dotted column) for 24 h. NF-κB activity of untreated controls was set at 100%. The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test. (e) We cultured MOLT4 stable transformants (mock and NICD) with the indicated concentrations of either bortezomib (upper panel) or mithramycin (lower panel) for 48 h and stained with annexin-V/PE in preparation for flow cytometry. The y axis shows cell viability calculated by the equation: 100–annexin-V positivity (%). The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test. (f) We treated MOLT4 stable transformants (mock and NICD) with the indicated drugs and evaluated cell proliferation using MTT assays after 72 h. The proliferation of untreated cells was set at 100%. The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test.
Combination of bortezomib and other anti-leukemic agents produces synergistic or additive cytotoxicity in T-ALL cells
It has been reported that constitutive activation of Notch signaling pathways underlies the resistance to anticancer drugs and ionizing irradiation in various malignancies.6, 7 We therefore hypothesized that bortezomib-induced suppression of Notch1 enhances the effects of other anti-leukemic agents. To test this hypothesis, we analyzed the combined effects of bortezomib and four key drugs for the treatment of T-ALL using the classical isobologram method.30 As shown in Figure 6a, the combination index of bortezomib and either DEX or ADM was far below 1.0, indicating a synergistic effect of the combination. The synergistic effect of bortezomib and ADM is fully compatible with the notion that Sp1-mediated p65 overexpression underlies ADM resistance in ALL cells.43 The combination index of bortezomib and either cytosine arabinoside or 4-hydroxycyclophospamide was around 1.0, suggesting an additive or sub-additive effect of the combination.
Synergistic or additive effects of bortezomib and other anti-leukemia drugs on T-ALL cells in vitro. (a) Jurkat and MOLT4 cells were treated with various anti-leukemia drugs in the absence or presence of bortezomib for 72 h to obtain dose-response curves of each combination. The combination index plots were generated by the CompuSyn software according to the method of Chou and Talalay. Combination index <1.0 means synergism of the two drugs. (b) MOLT4 cells were cultured in the absence (−) or presence (+) of 3 nM bortezomib and various doses (0–1000 nM) of DEX for 48 h, and subjected to immunoblotting for full-length Notch1 (FL), Notch1 TM, cleaved Notch1 (NICD) and GAPDH (internal control). (c) MOLT4 cells were cultured in the absence (−) or presence (+) of 3 nM bortezomib and various doses (0–30 nM) of doxorubicin (ADM) for 48 h, and subjected to immunoblotting for full-length Notch1 (FL), Notch1 TM, cleaved Notch1 (NICD) and GAPDH (internal control). (d) Mock- or NICD-carrying stable transformants of Jurkat and MOLT4 were treated with various anti-leukemia drugs in the absence or presence of bortezomib for 72 h to obtain dose-response curves of each combination. The combination index was calculated by the CompuSyn software according to the method of Chou and Talalay. Combination index <1.0 means synergism of the two drugs. The means±s.d. (bars) of three independent experiments are shown. Asterisks indicate P<0.05 by Student’s t-test.
Recently, Cialfi et al.9 reported that glucocorticoids induce transcriptional repression of the Notch1 gene through glucocorticoid-responsive elements. As glucocorticoid-responsive elements (–1995, –1075, –978 and –873) are located far upstream of major Sp1-binding sites (–392/–342), it is highly likely that bortezomib and glucocorticoids synergistically repress transcription of Notch1 in T-ALL cells. Immunoblot analysis showed that this was the case: DEX readily potentiated bortezomib-induced suppression of all forms of Notch1 (FL, TM and NICD) in a dose-dependent manner (Figure 6b). Similarly, ADM greatly enhanced bortezomib-induced downregulation of Notch1 especially NICD, although ADM alone did not affect the expression of Notch1 at all. These results suggest that NICD targeting underlies the synergistic effects of bortezomib and these agents. In support of this view, we demonstrated that NICD overexpression abolished the synergistic effects of bortezomib and either DEX or ADM in Jurkat and MOLT4 stable transformants (Figure 6d). Furthermore, the Sp1 inhibitor mithramycin was also synergistic with the same set of agents as bortezomib in T-ALL cells (Supplementary Figure 5). Taken together, bortezomib could elicit an anti-leukemic effect alone or in combination with conventional drugs on T-ALL cells by targeting the Sp1/Notch1 pathway.
Synergistic anti-leukemia effects of bortezomib and DEX in vivo
Finally, we validated therapeutic effects of bortezomib alone and in combination with DEX on T-ALL in vivo. For this purpose, we inoculated MOLT4 cells into NOD/SCID mice and treated them in four ways: the vehicle alone, DEX alone, bortezomib alone and the combination of both agents. As shown in Figure 7a, inoculated tumors rapidly grew in vehicle control mice between 7 and 14 days. Monotherapy with either DEX or bortezomib was able to retard tumor growth, but the effect of combination was striking with the almost complete disappearance of inoculated tumors in some recipients (Figures 7b–d). These observations confirm the results of in vitro experiments and further suggest that combination therapy including bortezomib is effective for T-ALL in clinical settings.
Synergistic effects of bortezomib and DEX on T-ALL cells in vivo. (a) We inoculated 1 × 107 MOLT4 cells into NOD/SCID mice and treated them in four ways: the vehicle alone (Control), DEX alone (DEX), bortezomib alone (Bor) and the combination of both agents (Bor+DEX). Treatments were started at day 0 when inoculated tumors were measurable. Tumor sizes were measured at the indicated time points to calculate tumor volume. (b) The y axis shows the average tumor volume at days 0, 14 and 21. The means±s.d. (bars) are shown (n=3–4). P-values were calculated by one-way analysis of variance (ANOVA) with the Tukey’s multiple comparison test (*P<0.05). (c) We inoculated 1 × 107 luciferase-expressing MOLT4 sublines into NOD/SCID mice and treated as described in (a). In vivo luciferase activity was measured by the IVIS Imaging System at day 14. (d) Quantitative data of in vivo bioluminescence imaging expressed as photon units (photons/s). The means±s.d. (bars) are shown (n=3). P-values were calculated by one-way ANOVA with the Tukey’s multiple comparison test (*P<0.05).
Discussion
In this study, we demonstrated that proteasome inhibitors effectively target constitutively active Notch1 to elicit cytotoxicity and increase drug sensitivity in T-ALL cells in vitro and in vivo. In contrast to GSIs, bortezomib is highly effective for T-ALL cells regardless of the presence of mutations at heterodimerization domains, because it downregulates the expression of Notch1 at a transcriptional level via degradation of Sp1, a pivotal transactivator of the Notch1 gene. This point is obviously advantageous over other Notch-targeting agents including GSIs and blocking antibodies against extracellular domains of Notch1. The reduction of NICD is not only cytotoxic to T-ALL cells but also enhances their sensitivity to other anti-leukemia drugs including glucocorticoids and doxorubicin. This is fully consistent with a previous finding that GSI-mediated inhibition of Notch signaling reverses glucocorticoid resistance in T-ALL cells.6 The safety and effectiveness of bortezomib were already established through long-term clinical experience.22 Taken together, this study provides a strong rationale for the inclusion of bortezomib in multidrug combination therapy for T-ALL patients to improve the treatment outcome and prognosis.
It is still possible that post-transcriptional mechanisms are also involved in Notch1 downregulation by proteasome inhibitors. It has been reported that Notch1 is cleaved into six fragments mainly by caspase-6 in dying T-lymphocytes.44 Moreover, Notch1 signaling is reported to be affected by lysosome activity.45 We therefore examined the effects of the caspase-6 inhibitor Z-VEID-fmk and the lysosome inhibitor chloroquine on Notch1 expression in bortezomib-treated T-ALL cells. We found no evidence of participation of these pathways in bortezomib-induced downregulation of Notch1 (data not shown). From these results, we provisionally concluded that bortezomib-induced decrease in Notch1 expression primarily occurs at a transcriptional level in T-ALL cells.
Downregulation of Sp1 seems to be responsible for transcriptional repression of Notch1 in bortezomib-treated T-ALL cells; however, the presence of additional mechanisms cannot be ruled out. Sp1 is known to undergo post-translational modifications such as phosphorylation, glycosylation, acetylation, sumoylation and ubiquitination.46 After sumoylation, Sp1 is ubiquitinated by the ubiquitin E3 ligase RNF4 and is degraded via ubiquitin-proteasome cascades. Bortezomib could inhibit this process to accumulate ubiquitinated Sp1, which perturbs the function of residual unmodified Sp1 in T-ALL cells. It is worth investigating that bortezomib inactivates the transcription of Sp1 target genes including Notch1 by perturbing the activator function of Sp1 via post-translational mechanisms.
Accumulating evidence indicates that Notch activates NF-κB pathways via multiple mechanisms. In CD3/CD28-stimulated murine T-lymphocytes, NICD directly binds to c-Rel and p50, and facilitates their nuclear retention by competing with IκBα.47 Forced expression of NICD reportedly induces nuclear translocation of p65, p50, c-Rel and RelB in hematopoietic progenitor cells.39 These observations are in line with our finding that NICD downregulation resulted in a decreased expression of p65 and p50 in the nuclei, although the underlying mechanisms are to be elucidated. It was demonstrated that NICD activates the IKK complex via CYLD downregulation25, 26 or direct interaction39 to trigger proteasome-dependent degradation of IκBα, leading to canonical NF-κB activation. It is possible that bortezomib reverses this process by targeting NICD to increase the abundance of IκBα, but our data did not support this view. Bortezomib was shown to activate IKKβ and decrease the abundance of IκBα, thereby increasing canonical NF-kB activity in myeloma cells;23 however, this was not reproducible in T-ALL cells. In T-ALL cells, bortezomib-mediated NICD downregulation perturbs IKKβ activity via destabilization of the IKK complex, but fails to increase the abundance of IκBα because of concomitant repression of the IκBα gene, a direct transcriptional target of NICD.39 In this regard, the synergistic effect of bortezomib and an IKK inhibitor in NICD-overexpressing cells is reasonable and offers the possibility of clinical translation of this combination in bortezomib refractory cases.48
The emerging question here is why downregulation of Sp1, which is widely expressed and activates the transcription of numerous genes including housekeepers, results in a selective repression of Notch1 with less effect on the transcription of other genes such as HDAC1 and GAPDH. A recent elegant study by Lovén et al.49 provides a good explanation. In cancer cells, the expression of genes harboring oncogenic mutations is driven by a unique element designated as super-enhancers, where a high level of multiple transcription factors and coactivators accumulate at a relatively wide distance. The super-enhancers are extremely susceptible to subtle changes in activator concentrations because of the high demand of cooperative binding and synergistic activation by their components. In case of T-ALL, the expression of its driver gene Notch1 might be sustained by Sp1 binding to super-enhancer at the region between –480 and –342, and thus, is extremely vulnerable to the loss of Sp1 by proteasome inhibitors. Nonetheless, addiction to Sp1 was demonstrated in cancer cells before the proposal of the concept of super-enhancers.24, 40, 50 As pointed out by Lovén et al.,49 super-enhancers are ideal therapeutic targets of various intractable cancers including MM and T-ALL.
References
Pui CH, Evans WE . Treatment of acute lymphoblastic leukemia. N Engl J Med 2006; 354: 166–178.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Callens C, Baleydier F, Lengline E, Abdelali RB, Petit A, Villarese P et al. Clinical impact of NOTCH1 and/or FBXW7 mutations, FLASH deletion, and TCR status in pediatric T-cell lymphoblastic lymphoma. J Clin Oncol 2012; 30: 1966–1973.
Jenkinson S, Koo K, Mansour MR, Goulden N, Vore A, Mitchell C et al. Impact of NOTCH1/FBXW7 mutations on outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on the MRC UKALL 2003 trial. Leukemia 2013; 27: 41–47.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 1996; 183: 2283–2291.
Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E et al. γ-Secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med 2009; 15: 50–58.
Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta 2010; 1806: 258–267.
Annino L, Vegna ML, Camera A, Specchia G, Visani G, Floritoni G et al. Treatment of adult acute lymphoblastic leukemia (ALL): long-term follow-up of the GIMEMA ALL 0288 randomized study. Blood 2002; 99: 863–871.
Cialfi S, Palermo R, Manca S, Checquolo S, Bellavia D, Pelullo M et al. Glucocorticoid sensitivity of T-cell lymphoblastic leukemia/lymphoma is associated with glucocorticoid receptor-mediated inhibition of Notch1 expression. Leukemia 2013; 27: 485–488.
Koch U, Radtke F . Notch in T-ALL: new players in a complex disease. Trends Immunol 2011; 32: 434–442.
Lobry C, Oh P, Aifantis I . Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med 2011; 208: 1931–1935.
Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC . Notch subunit heterodimerization and prevention of ligand independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol 2004; 24: 9265–9273.
Osipo C, Patel P, Rizzo P, Clementtz AG, Hao L, Golde TE et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a γ-secretase inhibitor. Oncogene 2008; 27: 5019–5032.
Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, Koga K et al. γ-Secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology 2008; 134: 131–144.
Meng RD, Shelton CC, Li Y-M, Qin L-X, Notterman D, Paty PB et al. γ-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res 2009; 69: 573–582.
Masuda S, Kumano K, Suzuki T, Tomita T, Iwatsubo T, Natsugari H et al. Dual antitumor mechanisms of Notch signaling inhibitor in a T-cell acute lymphoblastic leukemia xenograft model. Cancer Sci 2009; 100: 2444–2450.
Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol 2012; 30: 2307–2313.
Tolcher AW, Messersmith WA, Mikulski SM, Papadopoulos KP, Kwak EL, Gibbon DG et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol 2012; 30: 2348–2353.
Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem 2008; 283: 8046–8054.
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y et al. Therapeutic antibody targeting of individual Notch receptors. Nature 2010; 464: 1052–1057.
Aste-Amézaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One 2010; 5: e9094.
Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A et al. Proteasome inhibitors in multiple myeloma: 10 years later. Blood 2012; 120: 947–959.
Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K et al. Bortezomib induces canonical nuclear factor-κB activation in multiple myeloma cells. Blood 2009; 114: 1046–1052.
Kikuchi J, Wada T, Shimizu R, Izumi T, Akutsu M, Mitsunaga K et al. Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood 2010; 116: 406–417.
Espinosa L, Cathelin S, D'Altri T, Trimarchi T, Statnikov A, Guiu J et al. The Notch/Hes1 pathway sustains NF-κB activation through CYLD repression in T cell leukemia. Cancer Cell 2010; 18: 268–281.
D'Altri T, Gonzalez J, Aifantis I, Espinosa L, Bigas A . Hes1 expression and CYLD repression are essential events downstream of Notch1 in T-cell leukemia. Cell Cycle 2011; 10: 1031–1036.
Moreno DA, Scridel CA, Cortez MAA, de Paula Queiroz R, Valera ET, da Silva Silveira V et al. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol 2010; 150: 665–673.
Aldana-Masangkay GI, Rodriguez-Gonzalez A, Lin T, Ikeda AK, Hsieh Y-T, Kim Y-M et al. Tubacin suppresses proliferation and induces apoptosis of acute lymphoblastic leukemia cells. Leuk Lymphoma 2011; 52: 1544–1555.
Huang C, Hu X, Wang L, Lü S, Cheng H, Song X et al. Bortezomib suppresses the growth of leukemia cells with Notch1 overexpression in vivo and in vitro. Cancer Chemother Pharmacol 2012; 70: 801–809.
Chou TC . Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70: 440–446.
Wada T, Kikuchi J, Furukawa Y . Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep 2012; 13: 142–149.
Renard P, Ernest I, Houbion A, Art M, Le Calvez H, Raes M et al. Development of a sensitive multi-well colorimetric assay for active NFκB. Nucleic Acid Res 2001; 29: e21.
Kikuchi J, Shibayama N, Yamada S, Wada T, Nobuyoshi M, Izumi T et al. Homopiperazine derivatives as a novel class of proteasome inhibitors with a unique mode of proteasome binding. PLoS One 2013; 8: e61649.
Kikuchi J, Yamada S, Koyama D, Wada T, Nobuyoshi M, Izumi T et al. The novel orally active proteasome inhibitor K-7174 exerts anti-myeloma activity in vitro and in vivo by down-regulating the expression of class I histone deacetylases. J Biol Chem 2013; 288: 25593–25602.
Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 2010; 115: 2872–2881.
van Hamburg JP, de Bruijn MJW, Dingjan GM, Beverloo HB, Diepstraten H, Ling K-W et al. Cooperation of Gata3 c-Myc and Notch in malignant transformation of double positive thymocytes. Mol. Immunol 2008; 45: 3085–3095.
Giambra V, Jenkins CR, Wang H, Lam SH, Shevchuk OO, Nemirovsky O et al. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-θ and reactive oxygen species. Nat Med 2012; 18: 1693–1698.
Chang P-Y, Draheim K, Kelliher MA, Miyamoto S . NFKB1 is a direct target of the TAL1 oncoprotein in human T leukemia cells. Cancer Res 2006; 66: 6009–6013.
Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F et al. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat Med 2007; 13: 70–77.
Lambertini C, Pantano S, Dotto GP . Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal versus cancer-derived keratinocytes. PLoS One 2010; 5: e10369.
Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM . Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest 1991; 88: 1613–1621.
Deacon K, Onion D, Kumari R, Watson SA, Knox AJ . Elevated SP-1 transcription factor expression and activity drives basal and hypoxia-induced vascular endothelial growth factor (VEGF) expression in non-small cell lung cancer. J Biol Chem 2012; 287: 39967–39981.
Gu L, Findley HW, Zhou M . MDM2 induces NF-κB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood 2002; 99: 3367–3375.
Cohen LY, Bourbonnière M, Sabbagh L, Bouchard A, Chew T, Jeannequin P et al. Notch1 antiapoptotic activity is abrogated by caspase cleavage in dying T lymphocytes. Cell Death Differ 2005; 12: 243–254.
Zheng L, Saunders CA, Sorensen EB, Waxmonsky NC, Conner SD . Notch signaling from the endosome requires a conserved dileucine motif. Mol Biol Cell 2013; 24: 297–307.
Wang YT, Yang WB, Chang WC, Hung JJ . Interplay of posttranslational modifications in Sp1 mediates Sp1 stability during cell cycle progression. J Mol Biol 2011; 414: 1–14.
Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE et al. Notch1 augments NF-κB activity by facilitating its nuclear retention. EMBO J 2006; 25: 129–138.
Hideshima T, Chauhan D, Kiziltepe T, Ikeda H, Okawa Y, Podar K et al. Biologic sequelae of IκB kinase (IKK) inhibition in multiple myeloma: therapeutic implications. Blood 2009; 113: 5228–5236.
Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Fulciniti M, Amin S, Nanjappa P, Rodig S, Prabhala R, Li C et al. Significant biological role of Sp1 transactivation in multiple myeloma. Clin Cancer Res 2011; 17: 6500–6509.
Acknowledgements
This work was supported in part by the High-Tech Research Center Project for Private Universities: Matching Fund Subsidy from MEXT, a Grant-in-Aid for Scientific Research from JSPS and research grants from Japan Leukemia Research Fund, Takeda Science Foundation, the Naito Foundation, the Yasuda Medical Foundation, Mitsui Life Social Welfare Foundation and the Uehara Memorial Foundation (to JK and YF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
YF received research funding and honoraria from Janssen Pharmaceuticals K.K. and Novartis Pharmaceuticals Co. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Koyama, D., Kikuchi, J., Hiraoka, N. et al. Proteasome inhibitors exert cytotoxicity and increase chemosensitivity via transcriptional repression of Notch1 in T-cell acute lymphoblastic leukemia. Leukemia 28, 1216–1226 (2014). https://doi.org/10.1038/leu.2013.366
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.366
Keywords
This article is cited by
-
Phase 2 study of combination chemotherapy with bortezomib in children with relapsed and refractory acute lymphoblastic leukemia
International Journal of Hematology (2023)
-
Ciclopirox and bortezomib synergistically inhibits glioblastoma multiforme growth via simultaneously enhancing JNK/p38 MAPK and NF-κB signaling
Cell Death & Disease (2021)
-
Advances of target therapy on NOTCH1 signaling pathway in T-cell acute lymphoblastic leukemia
Experimental Hematology & Oncology (2020)
-
Differential antitumor activity of compounds targeting the ubiquitin-proteasome machinery in gastrointestinal stromal tumor (GIST) cells
Scientific Reports (2020)
-
Biology and Treatment Paradigms in T Cell Acute Lymphoblastic Leukemia in Older Adolescents and Adults
Current Treatment Options in Oncology (2020)