Abstract
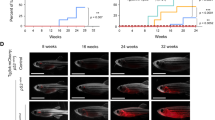
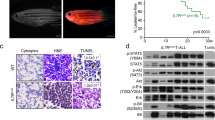
NOTCH1 pathway activation contributes to the pathogenesis of over 60% of T-cell acute lymphoblastic leukemia (T-ALL). While Notch is thought to exert the majority of its effects through transcriptional activation of Myc, it also likely has independent roles in T-ALL malignancy. Here, we utilized a zebrafish transgenic model of T-ALL, where Notch does not induce Myc transcription, to identify a novel Notch gene expression signature that is also found in human T-ALL and is regulated independently of Myc. Cross-species microarray comparisons between zebrafish and mammalian disease identified a common T-ALL gene signature, suggesting that conserved genetic pathways underlie T-ALL development. Functionally, Notch expression induced a significant expansion of pre-leukemic clones; however, a majority of these clones were not fully transformed and could not induce leukemia when transplanted into recipient animals. Limiting-dilution cell transplantation revealed that Notch signaling does not increase the overall frequency of leukemia-propagating cells (LPCs), either alone or in collaboration with Myc. Taken together, these data indicate that a primary role of Notch signaling in T-ALL is to expand a population of pre-malignant thymocytes, of which a subset acquire the necessary mutations to become fully transformed LPCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol 2003; 21: 3616–3622.
Armstrong F, Brunet de la Grange P, Gerby B, Rouyez M-C, Calvo J, Fontenay M et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood 2009; 113: 1730–1740.
Gerby B, Clappier E, Armstrong F, Deswarte C, Calvo J, Poglio S et al. Expression of CD34 and CD7 on human T-cell acute lymphoblastic leukemia discriminates functionally heterogeneous cell populations. Leukemia 2011; 25: 1249–1258.
Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011; 469: 362–367.
Clappier E, Gerby B, Sigaux F, Delord M, Touzri F, Hernandez L et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med 2011; 208: 653–661.
Lee S-Y, Kumano K, Masuda S, Hangaishi A, Takita J, Nakazaki K et al. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia 2005; 19: 1841–1843.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS . Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol 2000; 20: 7505–7515.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 1996; 183: 2283–2291.
Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay A-S et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood 2010; 116: 5443–5454.
O’Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH et al. Activating Notch1 mutations in mouse models of T-ALL. Blood 2006; 107: 781–785.
Treanor LM, Volanakis EJ, Zhou S, Lu T, Sherr CJ, Sorrentino BP . Functional interactions between Lmo2, the Arf tumor suppressor, and Notch1 in murine T-cell malignancies. Blood 2011; 117: 5453–5462.
Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA . Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood 2011; 118: 1579–1590.
Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H . Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med 2008; 205: 2851–2861.
McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 2010; 327: 879–883.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA 2006; 103: 18261–18266.
Sharma VM, Draheim KM, Kelliher MA . The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle 2007; 6: 927–930.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 2006; 20: 2096–2109.
Knoepfler PS . Why myc? an unexpected ingredient in the stem cell cocktail. Cell Stem Cell 2008; 2: 18–21.
Demarest RM, Dahmane N, Capobianco AJ . Notch is oncogenic dominant in T-cell acute lymphoblastic leukemia. Blood 2011; 117: 2901–2909.
Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev 1996; 10: 1930–1944.
Pajcini KV, Speck NA, Pear WS . Notch signaling in mammalian hematopoietic stem cells. Leukemia 2011; 25: 1525–1532.
Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003; 299: 887–890.
Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD . NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 2007; 21: 462–471.
Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT . Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA 2005; 102: 6068–6073.
Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 2010; 18: 353–366.
Blackburn JS, Liu S, Raimondi AR, Ignatius MS, Salthouse CD, Langenau DM . High-throughput imaging of adult fluorescent zebrafish with an LED fluorescence macroscope. Nat Protoc 2011; 6: 229–241.
Assaf C, Hummel M, Dippel E, Goerdt S, Müller HH, Anagnostopoulos I et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood 2000; 96: 640–646.
Assaf C, Hummel M, Steinhoff M, Geilen CC, Orawa H, Stein H et al. Early TCR-beta and TCR-gamma PCR detection of T-cell clonality indicates minimal tumor disease in lymph nodes of cutaneous T-cell lymphoma: diagnostic and prognostic implications. Blood 2005; 105: 503–510.
Blackburn JS, Liu S, Langenau DM . Quantifying the frequency of tumor-propagating cells using limiting dilution cell transplantation in syngeneic zebrafish. J Vis Exp 2011; 53: 1–5, e2790.
Smith ACH, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV . High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood 2010; 115: 3296–3303.
Hu Y, Smyth GK . ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009; 347: 70–78.
Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 2007; 21: 1382–1395.
De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med 2010; 16: 1321–1327.
Guo Z, Dose M, Kovalovsky D, Chang R, O’Neil J, Look AT et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood 2007; 109: 5463–5472.
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, JGCAM Buijs-Gladdines, STCJM Peters et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 2009; 10: 125–134.
Langenau DM, Keefe MD, Storer NY, Jette CA, Smith ACH, Ceol CJ et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 2008; 27: 4242–4248.
Ferrando AA, Look AT . Gene expression profiling in T-cell acute lymphoblastic leukemia. Semin Hematol 2003; 40: 274–280.
Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC et al. Direct inhibition of the NOTCH transcription factor complex. Nature 2009; 462: 182–188.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357.
Mori S, Rempel RE, Chang JT, Yao G, Lagoo AS, Potti A et al. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Emicro-myc model and human diffuse large B-cell lymphoma. Cancer Res 2008; 68: 8525–8534.
Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 2007; 11: 349–360.
Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JPP . Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol 2008; 143: 153–168.
Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hébert J, Perreault C et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev 2010; 24: 1093–1105.
Wendorff AA, Koch U, Wunderlich FT, Wirth S, Dubey C, Brüning JC et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 2010; 33: 671–684.
Magri M, Yatim A, Benne C, Balbo M, Henry A, Serraf A et al. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur J Immunol 2009; 39: 1231–1240.
Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet 2007; 39: 593–595.
Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood 2003; 102: 262–268.
Acknowledgements
We thank Dr Ravi Mylvaganam for expert advice and assistance with cell sorting, Eric Stone and Marcellino Pena for excellent animal husbandry, and Dr Finola Moore and Tommy Jones help with data interpretation and critical review of the manuscript. JSB is supported by NIH 5T32CA09126-26. DML is supported by an American Cancer Society Research Scholar Grant, the Leukemia Research Foundation, the Alex Lemonade Stand Foundation, the Harvard Stem Cell Institute, and NIH K01AR05562190.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Blackburn, J., Liu, S., Raiser, D. et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia 26, 2069–2078 (2012). https://doi.org/10.1038/leu.2012.116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.116
Keywords
This article is cited by
-
IRF4 drives clonal evolution and lineage choice in a zebrafish model of T-cell lymphoma
Nature Communications (2022)
-
Mutant IL7R collaborates with MYC to induce T-cell acute lymphoblastic leukemia
Leukemia (2022)
-
PRL3 enhances T-cell acute lymphoblastic leukemia growth through suppressing T-cell signaling pathways and apoptosis
Leukemia (2021)
-
Protein tyrosine phosphatase 4A3 (PTP4A3/PRL-3) drives migration and progression of T-cell acute lymphoblastic leukemia in vitro and in vivo
Oncogenesis (2020)
-
Simultaneous B and T cell acute lymphoblastic leukemias in zebrafish driven by transgenic MYC: implications for oncogenesis and lymphopoiesis
Leukemia (2019)