Abstract
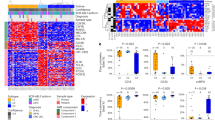
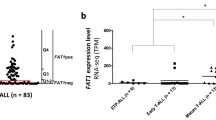
Improved survival of patients with acute lymphoblastic leukemia (ALL) has emerged from identifying new prognostic markers; however, 20% of children still suffer recurrence. Previously, the altered expression of Fat1 cadherin has been implicated in a number of solid tumors. In this report, in vitro analysis shows that Fat1 protein is expressed by a range of leukemia cell lines, but not by normal peripheral blood (PB) and bone marrow (BM) cells from healthy donors. In silico analysis of expression of array data from clinical leukemias found significant levels of Fat1 transcript in 11% of acute myeloid leukemia, 29% and 63% of ALL of B and T lineages, respectively, and little or no transcript present in normal PB or BM. Furthermore, in two independent studies of matched diagnosis–relapse of precursor B-cell (preB) ALL pediatric samples (n=32 and n=27), the level of Fat1 mRNA expression was prognostic at the time of diagnosis. High Fat1 mRNA expression was predictive of shorter relapse-free and overall survival, independent of other traditional prognostic markers, including white blood cell count, sex and age. The data presented demonstrate that Fat1 expression in preB-ALL has a role in the emergence of relapse and could provide a suitable therapeutic target in high-risk preB-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood 2008; 111: 776–784.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764.
Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 2009; 114: 2159–2167.
Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 2007; 21: 1258–1266.
Strefford JC, Worley H, Barber K, Wright S, Stewart AR, Robinson HM et al. Genome complexity in acute lymphoblastic leukemia is revealed by array-based comparative genomic hybridization. Oncogene 2007; 26: 4306–4318.
Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 2010; 24: 1258–1264.
Borowitz MJ, Pullen DJ, Winick N, Martin PL, Bowman WP, Camitta B . Comparison of diagnostic and relapse flow cytometry phenotypes in childhood acute lymphoblastic leukemia: implications for residual disease detection: a report from the children's oncology group. Cytometry B Clin Cytom 2005; 68: 18–24.
Henderson MJ, Choi S, Beesley AH, Sutton R, Venn NC, Marshall GM et al. Mechanism of relapse in pediatric acute lymphoblastic leukemia. Cell Cycle 2008; 7: 1315–1320.
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008; 322: 1377–1380.
Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011; 469: 356–361.
Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011; 469: 362–367.
Collins-Underwood JR, Mullighan CG . Genomic profiling of high-risk acute lymphoblastic leukemia. Leukemia 2010; 24: 1676–1685.
Dunne J, Hanby AM, Poulsom R, Jones TA, Sheer D, Chin WG et al. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 1995; 30: 207–223.
Ponassi M, Jacques TS, Ciani L, ffrench Constant C . Expression of the rat homologue of the Drosophila fat tumour suppressor gene. Mech Dev 1999; 80: 207–212.
Cox B, Hadjantonakis AK, Collins JE, Magee AI . Cloning and expression throughout mouse development of mfat1, a homologue of the Drosophila tumour suppressor gene fat. Dev Dyn 2000; 217: 233–240.
Down M, Power M, Smith SI, Ralston K, Spanevello M, Burns GF et al. Cloning and expression of the large zebrafish protocadherin gene, Fat. Gene Expr Patterns 2005; 5: 483–490.
Settakorn J, Kaewpila N, Burns GF, Leong AS . FAT, E-cadherin, beta catenin, HER 2/neu, Ki67 immuno-expression, and histological grade in intrahepatic cholangiocarcinoma. J Clin Pathol 2005; 58: 1249–1254.
Kwaepila N, Burns G, Leong AS . Immunohistological localisation of human FAT1 (hFAT) protein in 326 breast cancers. Does this adhesion molecule have a role in pathogenesis? Pathology 2006; 38: 125–131.
Sadeqzadeh E, de Bock CE, Zhang XD, Shipman KL, Scott NM, Song C et al. Dual processing of Fat1 cadherin by human melanoma cells generates distinct protein products. J Biol Chem 2011; 286: 28181–28191.
Katoh Y, Katoh M . Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med 2006; 18: 523–528.
Chetcuti A, Aktas S, Mackie N, Ulger C, Toruner G, Alkan M et al. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr Blood Cancer 2011; 57: 950–957.
Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011; 144: 296–309.
Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM et al. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol 2005; 3: e254.
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004; 350: 1617–1628.
Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. Blood 2006; 108: 711–717.
Staal FJ, de Ridder D, Szczepanski T, Schonewille T, van der Linden EC, van Wering ER et al. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia 2010; 24: 491–499.
Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia 2008; 22: 2142–2150.
Smith M, Arthur D, Camitta B, Carroll AJ, Crist W, Gaynon P et al. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol 1996; 14: 18–24.
Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2011; 117: 6267–6276.
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463: 899–905.
Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 2007; 447: 966–971.
Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res 2008; 14: 4572–4583.
Staal FJ, van der Burg M, Wessels LF, Barendregt BH, Baert MR, van den Burg CM et al. DNA microarrays for comparison of gene expression profiles between diagnosis and relapse in precursor-B acute lymphoblastic leukemia: choice of technique and purification influence the identification of potential diagnostic markers. Leukemia 2003; 17: 1324–1332.
Greaves M . Cancer stem cells: back to Darwin? Semin Cancer Biol 2010; 20: 65–70.
Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 2010; 10: 4874–4884.
Hawkins ED, Russell SM . Upsides and downsides to polarity and asymmetric cell division in leukemia. Oncogene 2008; 27: 7003–7017.
Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B et al. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A 2009; 106: 8579–8584.
Grusche FA, Richardson HE, Harvey KF . Upstream regulation of the hippo size control pathway. Curr Biol 2010; 20: R574–R582.
Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell 2010; 18: 579–591.
Tanoue T, Takeichi M . Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J Cell Biol 2004; 165: 517–528.
Ge X, Wang X . Role of Wnt canonical pathway in hematological malignancies. J Hematol Oncol 2010; 3: 33.
Nygren MK, Dosen-Dahl G, Stubberud H, Walchli S, Munthe E, Rian E . beta-catenin is involved in N-cadherin-dependent adhesion, but not in canonical Wnt signaling in E2A-PBX1-positive B acute lymphoblastic leukemia cells. Exp Hematol 2009; 37: 225–233.
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 2009; 10: 125–134.
Li Z, Zhang W, Wu M, Zhu S, Gao C, Sun L et al. Gene expression-based classification and regulatory networks of pediatric acute lymphoblastic leukemia. Blood 2009; 114: 4486–4493.
Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood 2003; 102: 2951–2959.
Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002; 1: 133–143.
Raimondi SC, Behm FG, Roberson PK, Williams DL, Pui CH, Crist WM et al. Cytogenetics of pre-B-cell acute lymphoblastic leukemia with emphasis on prognostic implications of the t(1;19). J Clin Oncol 1990; 8: 1380–1388.
Vey N, Thomas X, Picard C, Kovascovicz T, Charin C, Cayuela JM et al. Allogeneic stem cell transplantation improves the outcome of adults with t(1;19)/E2A-PBX1 and t(4;11)/MLL-AF4 positive B-cell acute lymphoblastic leukemia: results of the prospective multicenter LALA-94 study. Leukemia 2006; 20: 2155–2161.
Jeha S, Pei D, Raimondi SC, Onciu M, Campana D, Cheng C et al. Increased risk for CNS relapse in pre-B cell leukemia with the t(1;19)/TCF3-PBX1. Leukemia 2009; 23: 1406–1409.
Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009; 360: 2730–2741.
Ciani L, Patel A, Allen ND, ffrench-Constant C . Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol 2003; 23: 3575–3582.
Smith TG, Van Hateren N, Tickle C, Wilson SA . The expression of Fat-1 cadherin during chick limb development. Int J Dev Biol 2007; 51: 173–176.
Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE . The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J Cell Biol 2006; 173: 417–429.
Acknowledgements
We are grateful to Dheepa Bhojwani who made clinical outcome data available. We thank Professor Murray Norris for his comments and critical review of the manuscript. This work was supported by a project grant from the National Health and Medical Research Council, Australia (455531), Hunter Medical Research Institute (HMRI-09-23 kindly sponsored by Gallerie Fine Jewellery) and the Leukaemia Foundation Australia. RFT was awarded a career development fellowship by the Cancer Institute NSW. AA is in receipt of the Arrow Bone Marrow Transplant Foundation Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
de Bock, C., Ardjmand, A., Molloy, T. et al. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis–relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia 26, 918–926 (2012). https://doi.org/10.1038/leu.2011.319
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2011.319
Keywords
This article is cited by
-
FAT1 expression in T-cell acute lymphoblastic leukemia (T-ALL) modulates proliferation and WNT signaling
Scientific Reports (2023)
-
The diverse functions of FAT1 in cancer progression: good, bad, or ugly?
Journal of Experimental & Clinical Cancer Research (2022)
-
Detailed characterization of the transcriptome of single B cells in mantle cell lymphoma suggesting a potential use for SOX4
Scientific Reports (2021)
-
NFкB is a critical transcriptional regulator of atypical cadherin FAT1 in glioma
BMC Cancer (2020)
-
Detecting qualitative changes in biological systems
Scientific Reports (2020)