Abstract
Invasive aspergillosis is an emerging threat to public health due to the increasing use of immune suppressive drugs and the emergence of resistance against antifungal drugs. To deal with this threat, research on experimental disease models provides insight into the pathogenesis of infections caused by susceptible and resistant Aspergillus strains and by assessing their response to antifungal drugs. However, standard techniques used to evaluate infection in a preclinical setting are severely limited by their invasive character, thereby precluding evaluation of disease extent and therapy effects in the same animal. To enable non-invasive, longitudinal monitoring of invasive pulmonary aspergillosis in mice, we optimized computed tomography (CT) and magnetic resonance imaging (MRI) techniques for daily follow-up of neutropenic BALB/c mice intranasally infected with A. fumigatus spores. Based on the images, lung parameters (signal intensity, lung tissue volume and total lung volume) were quantified to obtain objective information on disease onset, progression and extent for each animal individually. Fungal lung lesions present in infected animals were successfully visualized and quantified by both CT and MRI. By using an advanced MR pulse sequence with ultrashort echo times, pathological changes within the infected lung became visually and quantitatively detectable at earlier disease stages, thereby providing valuable information on disease onset and progression with high sensitivity. In conclusion, these non-invasive imaging techniques prove to be valuable tools for the longitudinal evaluation of dynamic disease-related changes and differences in disease severity in individual animals that might be readily applied for rapid and cost-efficient drug screening in preclinical models in vivo.
Similar content being viewed by others
Main
Aspergillus fumigatus is clinically the most important Aspergillus species that can cause a spectrum of diseases in humans, described by the term aspergillosis.1, 2 The main route of infection in humans is via inhalation of fungal spores.3, 4 In healthy people, this rarely results in the development of sino-pulmonary infection, because the fungal conidia are rapidly cleared from the airways by the innate immune system.5 However, certain risk factors are associated with an increased chance of developing allergic broncho-pulmonary aspergillosis, aspergilloma or invasive aspergillosis.3 Invasive pulmonary aspergillosis (IPA) mainly affects immunocompromised individuals such as solid organ transplant recipients and patients with hematological malignancies, including hematopoietic stem cell transplant recipients. This disease is associated with significant morbidity and carries a crude mortality rate of up to 30–40% in some risk groups.6, 7, 8 In addition, a growing number of azole-resistant A. fumigatus strains has been identified in Europe over the past decade, contributing significantly to treatment failure.9, 10, 11, 12, 13
Preclinical in vivo studies are indispensable for gaining a better understanding of the pathogenesis of infections caused by both azole-susceptible and -resistant strains and for assessing their response to antifungal drugs. However, the gold standard techniques used for evaluation of infections in small animal models, such as fungal load quantification by colony-forming unit (CFU) counts, galactomannan (GM) antigen detection and microscopy of tissue sections, have limitations. Histological analysis of lung tissue provides valuable insight into cellular interactions, but it requires killing numerous animals at each time point, is very labor intensive and only provides a snapshot image of the infection. The number of viable fungal cells within lung tissue (CFU counts) is determined by manual counting of colonies after homogenizing the tissue and plating dilution series. In addition to the invasiveness of this technique, it is difficult to quantify colonies in case of IPA due to hyphae formation. Therefore, CFU counts do not provide reliable quantification of disease severity. In recent years, GM detection has become more important as a biomarker for diagnosis of invasive aspergillosis in both clinical and preclinical settings.2 Nonetheless, a substantial amount of blood or broncho-alveolar lavage (BAL) fluid is needed to perform the test, which hampers repeated measurements in small animals such as mice.
There is a strong need for alternative techniques to enable the dynamic follow-up of disease development and progression in infected animals, especially in preclinical drug screening studies. In most of these studies, antifungal treatment is initiated soon after infection without knowing the exact disease state in individual animals, in spite of the highly variable response upon spore administration.14, 15 By dynamically monitoring the course of disease using non-invasive imaging techniques, animals that did not develop infection could be removed from the study. Furthermore, treatment initiation could potentially be adjusted for individual animals, given the use of an appropriate experimental set-up, and the time-dependent effect of the antifungal can be evaluated in great detail, thereby providing valuable information on drug efficacy.
Some studies have evaluated the potential of optical imaging techniques, such as bioluminescence imaging (BLI), to visualize pulmonary aspergillosis in experimental animal models.16, 17, 18, 19 BLI appeared to be suitable to investigate the effectiveness of antifungal compounds in vitro but proved to be very challenging to detect infection in vivo, possibly owing to problems with substrate availability.20 Nuclear imaging techniques, such as positron emission tomography (PET) and single-photon emission computed tomography (SPECT), have also been used for detecting pulmonary aspergillosis in rodents by using radioactive probes.21, 22 As the use of radioactive tracers requires their production in cyclotrons, PET and SPECT may not always be the most practical methods for a preclinical setting. Also, the selectivity of PET tracers, such as 68Ga-siderophores and 89Zr-oxalate, for detecting Aspergillus infections has yet to be investigated in more detail.21, 23
Computed tomography (CT) is highly suitable for lung applications because it produces high-resolution 3D images (<100 μm) with an excellent air–tissue contrast.24, 25 The value of this imaging technique has been studied in disease models of lung fibrosis, lung emphysema and lung neoplasms.26, 27, 28, 29, 30 Nevertheless, this technique has not been used for the assessment of invasive aspergillosis in small animals. Magnetic resonance imaging (MRI) provides images with good spatial resolution and excellent soft tissue contrast, thereby creating the potential to distinguish between different pathological processes occurring within the lung, such as inflammation or necrosis. However, MRI of the lung is challenging owing to the lack of detectable protons in air-filled spaces and potential artifacts between air–tissue interfaces.31, 32 Despite these challenges, the potential of MRI has motivated several groups to report the successful use of 1H-MRI for preclinical imaging of lung fibrosis and lung neoplasms.33, 34, 35 Recently, more advanced MR sequences were optimized to overcome sensitivity problems of lung MRI, in particular MRI using ultrashort echo times (UTEs) or zero echo times. These novel MR sequences were successfully used for lung fibrosis and emphysema imaging.31, 36, 37, 38 Despite the obvious advantages of both imaging techniques, CT and MRI have rarely been used for the in vivo investigation of pulmonary fungal infectious diseases models. The aim of this study was to evaluate the potential of CT and MR imaging techniques for the longitudinal, non-invasive assessment of disease development and progression in mice suffering from IPA. Multiple potential biomarkers were quantified to obtain objective information on the course of disease in individual animals and compared with gold standard techniques for validation.
MATERIALS AND METHODS
Fungal Strains
Two A. fumigatus strains were used in this study. The ASFU 1731 strain was originally cultured from a patient suffering from invasive aspergillosis (UZ Leuven, Belgium) and the FGSC A1258 strain was obtained from the fungal genetic stock centre (Kansas City, MO, USA). The strains were cultured for 3 days at 42 °C on diluted Sabouraud agar, containing 10% of the dextrose and peptone concentrations used in regular sabouraud agar. Conidia were harvested by flooding the agar with saline–0.1% Tween 80 (Sigma-Aldrich, Diegem, Belgium) and gently scraping the surface. The collected suspension was shaken vigorously for 5 min, and spores were counted with a Neubauer hemocytometer. The suspension was diluted to a final concentration of 2.5 × 107 or 1.0 × 107 spores/ml.
Mouse Model
All animal experiments were carried out in compliance with national and European regulations and were approved by the animal ethics committee of KU Leuven. Ten-week-old male BALB/c mice (Janvier, Le Genest, France) were rendered neutropenic by intraperitoneal injections of cyclophosphamide (150 mg/kg) on days 4 and 1 prior to instillation with fungal spores or saline. The animals infected with the low inoculum (2 × 105 A. fumigatus spores) received an additional intraperitoneal injection of cyclophosphamide (150 mg/kg) on day 2 after infection to retain neutropenia. A broad-spectrum antibioticum (Baytril, 5–8 mg/kg/day) was added to the drinking water to prevent bacterial infection. All animals were intranasally instilled on day 0 with either 5 × 105 A. fumigatus spores in 20 μl (n=10 for ASFU 1731 strain or n=8 for FGSC A1258 strain), 2 × 105 A. fumigatus spores in 20 μl (n=7 for FGSC A1258 strain) or saline (20 μl, n=5). Directly after instillation, the animals were positioned upright until normal breathing was resumed. Starting from day 1 after infection, all animals were imaged daily by MRI and CT. During image acquisition, the mice were anesthetized with 1.5–2% isoflurane (Abbott Laboratories, Queenborough, UK) in 100% oxygen (O2) administered via a nasal cone. Body weight, posture and body temperature were monitored daily. The animals were killed when humane end points were reached, including a strong loss in body weight (>25%), lethargy and labored breathing.
Computed Tomography
Retrospectively respiratory-gated lung CT data were acquired on a dedicated small animal CT scanner (SkyScan 1076, Bruker CT, Kontich, Belgium). The following scan parameters were used: 50 kVp X-ray source, 0.5 mm aluminium filter, 180 μA current, 120 ms exposure time per projection, 23 × 35 mm2 field of view (FOV) covering the lung, 9 projections per position with 0.7° increments and a total angle of 180°. The total acquisition time per scan was approximately 13 min, resulting in a reconstructed 3D data set with an isotropic voxel size of (35 × 35 × 35) μm3.
Magnetic Resonance Imaging
Images were acquired on a 9.4 Tesla Biospec small animal MRI scanner (20 cm horizontal bore size; Bruker BioSpin, Ettlingen, Germany) combined with a gradient insert (maximal field strength of 1200 mT/m) and a 3.5-cm quadrature resonator. Body temperature and breathing rate were continuously monitored and kept at physiological values during the scans by using a physiological monitoring system (SAII, Stony Brook, NY, USA). Three different MRI protocols were evaluated in this study, including a 2D and 3D self-gated (IntraGate, Paravision 5.1, Bruker) fast low angle shot (FLASH) gradient echo pulse sequence and a 3D prospectively respiratory-gated UTE pulse sequence, with the following parameters: Self-gated 2D-FLASH37: TR/TE 30/1.26 ms, 17° flip angle (FA), 5 slices covering the lung with a slice thickness of 1 mm and inter-slice gap of 0.5 mm, 4 cm isotropic FOV, 256 × 256 matrix, in-plane resolution of 156 × 156 μm2, 80 repetitions and a 1-cm wide navigator slab, excited with a 0.8-ms sinc10H pulse with a 1.5° FA, total acquisition time of 10 min. For reconstruction, 70% of the respiration and ECG period was used. Self-gated 3D-FLASH pulse sequence: 0.99 ms TE, 15 ms TR, 17° FA, 3 cm isotropic FOV, 160 × 160 × 160 matrix, 187 μm isotropic resolution, 6 repetitions resulting in a total acquisition time of 22 min. For reconstructing the images, 70% of the respiration and ECG period was used. Prospectively gated 3D-UTE pulse sequence: 0.03 ms TE, 15 ms TR, 5° FA, 3.5 cm isotropic FOV, 128 × 128 × 128 matrix, 273 μm isotropic resolution, and a total acquisition time of 18 min.
Image Analysis
The CT data was retrospectively gated, reconstructed and processed by using the software from the manufacturer (Tsort version 1.1.3.0, Nrecon version 1.6.8.0, Dataviewer version 1.5.0.2, CTAn version 1.14.4.1; Bruker, Ettlingen, Germany) as validated and described before.26, 39 Retrospective respiratory gating resulted in four data sets per scan, corresponding to different phases of the breathing cycle. The data set obtained during the end expiratory phase of the breathing was used for further analysis. The signal intensity, representing the X-ray attenuation, and total lung volume were quantified based on a volume of interest (VOI) covering the entire lung (Figure 1a, dashed line). This VOI was composed of multiple regions of interest (ROI) that were manually delineated on each coronal CT image slice, excluding the heart and main pulmonary vessels.37 The signal intensity was determined by selecting the complete histogram of brightness distribution (minimum threshold 0; maximum threshold 255) and calculating the mean brightness value. The lung tissue volume was determined by setting a fixed threshold (absolute value of 55) to select all voxels with a signal intensity above this threshold within the VOI.
CT of mouse lungs infected with Aspergillus fumigatus. (a) Coronal CT images of representative mouse lungs after intranasal instillation with saline (control, top row) or A. fumigatus FGSC A1258 (infection, bottom row). A manually defined ROI is shown by the dashed line. (b) Graph representing the cumulative image score derived from the CT images of mice instilled with saline or A. fumigatus spores (ASFU 1731 or FGSC A1258 strain). (c–e) Graphs representing the signal intensity, lung tissue volume and total lung volume quantified from the CT lung images. Error bars represent s.d. of multiple mice (saline: n=5, ASFU 1731: n=7–10, FGSC A1258: n=7–8). *P<0.05; ***P<0.001; ****P<0.0001.
The 2D-FLASH MR images were retrospectively gated and reconstructed by using software from the manufacturer (Paravision version 5.1; Bruker, Ettlingen, Germany). Image data obtained during the end expiratory phase of the breathing was used for further processing using the ImageJ software (version 1.6.0_2; NIH, Bethesda, MD, USA). The signal intensity, averaged over the entire lung, was quantified based on a VOI covering the complete lung by manually delineating a ROI on each image slice, excluding the heart and main pulmonary vessels. To correct for inter-scan variations in overall intensity, the calculated signal intensity was normalized to a reference signal intensity measured by placing a second circular ROI in chest muscle tissue, as described before.37 The total lung volume was calculated by manually delineating the lungs on each slice, including the heart and main blood vessels, and multiplying the resulting values with the sum of the slice thickness and inter-slice distance.
The 3D-UTE and 3D-FLASH MR images, obtained during the end expiratory phase by prospective gating, were processed similarly to the 2D-FLASH images by using an in-house written Mevislab module (version 2.6.1; Mevislab Medical Solutions and Fraunhofer MEVIS, Bremen, Germany). To calculate the lung tissue volume, an absolute value was used as fixed threshold for the 3D-UTE images to select all voxels with a signal intensity above this threshold. A variable threshold was used for the 3D-FLASH images, based on the receiver gain value of the individual scans, to correct for inter-scan variations in this parameter.
To semiquantitatively describe visual observations, a previously described scoring system was adapted and applied to determine the cumulative image score.40, 41, 42 At day 1 after infection, an initial score of zero was assigned to the CT/MRI lung data set of each individual animal. Subsequently, four coronal image slices were selected from each data set at four different, predefined positions within the lungs. Corresponding lung slices of the same animal acquired on day 2 after infection were visually compared with the lung slices selected at day 1. Subsequently, a value of −0.25, 0 or +0.25 was assigned to each of the selected slices depending on a respective worsening, stabilization or improvement of the pulmonary damage based on the observable changes in lung tissue volume. The resulting values for the four slices were added to obtain the cumulative image score at day 2. Based on the same principle, the images of day 3 were scored by comparing them to the images of day 2 and the obtained values were added to the cumulative image score of day 2, resulting in the cumulative image score at day 3, etc. Image scoring was performed by two independent, blinded researchers, yielding similar results (data not shown).
GM Assay and Fungal Load Quantification
After the last imaging time point, all animals were killed by an overdose of pentobarbital (Nembutal, CEVA Santé Animale, Diegem, Belgium). Blood was collected, centrifuged for 10 min at 1500 g, and the supernatant was used for further analysis. BAL was performed by inserting a catheter (22-gauge) into the trachea, followed by the instillation of 0.7 ml saline and aspiration of the fluid after 10 s. This procedure was repeated twice. GM levels in BAL fluid and serum samples were determined by using an Aspergillus enzyme immunoassay (Platelia, Bio-Rad, Marnes-La-Coquette, France). After collection of BAL fluid, the right lung lobes were aseptically removed for fungal load quantification by CFU counting. The lungs were weighed and homogenized in 600 μl phosphate-buffered saline. Ten-fold dilution series were prepared and plated on Sabouraud agar, followed by 3 days of incubation at 30 °C and manual counting of CFUs.
Histological Analysis
The left lung was inflated with 0.5 ml 10% formalin by inserting a 22-gauge catheter in the left main bronchus, isolated, postfixed (24 h in 10% formalin) and subsequently embedded in paraffin. The entire lung was sectioned (5 μm) and stained with periodic acid-Schiff (PAS), which stains fungi red.
Statistics
The data were analyzed using Prism (version 5.04; GraphPad software, San Diego, CA, USA). Repeated-measures ANOVA with a Bonferroni posttest was used to investigate changes in imaging-derived parameters over time. First, the last time point was completely excluded from the test to be able to investigate potential differences between the infected and non-infected animals on days 1, 2 and 3. Second, all animals with missing values on day 4 were excluded from the test solely to investigate potential differences between the infected and non-infected animals on day 4. Student’s t-test was used to evaluate differences in GM index and CFU counts between the different groups. Differences were considered statistically significant if P-value<0.05. In the figures, the data are represented as mean±s.d.
RESULTS
CT Enables Longitudinal Visualization and Quantification of IPA
To investigate the possibility to use optimized in vivo CT for monitoring of IPA, mice were infected with two different A. fumigatus strains and scanned daily until 4 days after infection. Lesions developing within the lungs of infected animals appeared as hyperintense patches against the darker background of normal, aerated lung tissue owing to absorption of the X-rays (Figure 1a, white arrows). In contrast, no changes were observed in the lungs of control animals during the course of the experiment (Figure 1a, upper row). Semiquantitative assessment of disease progression showed a significant increase in the cumulative score on days 3 and 4 after infection for both Aspergillus strains (Figure 1b). In addition, the signal intensity in the lungs showed a modest increase on day 3, becoming more prominent on day 4 (Figure 1c). This increase in signal intensity reached significance on day 4 for the group infected with the FGSC A1258 strain. For both Aspergillus strains, large s.ds. were observed on the last time points, illustrating a large inter-animal variation in advanced stages of infection. The lung tissue volume, which includes all lesions, showed for both strains a significant increase on days 3 and 4 after infection (Figure 1d). Markedly, the total lung volume showed a steady increase over time that reached significance on day 4 for both strains (Figure 1e). No significant differences could be detected between the two groups infected with different A. fumigatus strains. These results show that CT provides both visual and quantitative information to evaluate the course of disease in individual animals.
FLASH MRI Enables the Visual and Quantitative Assessment of Advanced IPA
In order to investigate the capability of MRI to non-invasively monitor disease progression in mice suffering from IPA, we first evaluated a self-gated gradient-echo sequence (2D-IntraGate-FLASH) to image mice infected with 5 × 105 conidia. On the acquired 2D-FLASH images, the lungs appeared hypointense, whereas the heart and main blood vessels appeared hyperintense (Figure 2a). Compared with the control animals, additional hyperintense signals appeared within the lungs of infected animals on day 4 (Figure 2a). In correspondence with this observation, the signal intensity quantified from these images remained stable for the first 3 days for both Aspergillus strains, only to rise significantly on day 4 (Figure 2b). Upon visual inspection of the images, the size of the lungs appeared to be increasing from day 3 after infection (Figure 2a, bottom row). In correspondence with this observation, quantification of the total lung volumes showed an increasing trend for both Aspergillus strains (Figure 2c). On the contrary, no changes were detected in the lungs of uninfected animals. These results indicate that 2D-FLASH MRI can be used as an imaging technique for longitudinal visualization and quantification of IPA in mice.
2D-FLASH MRI (self-gated) of Aspergillus fumigatus-infected mouse lungs. (a) Representative coronal 2D-FLASH images of the lungs after instillation with saline (control, top row) or A. fumigatus FGSC A1258 (infection, bottom row). Dashed line depicts a manually defined ROI. (b, c) Graphs representing the normalized signal intensity and total lung volume quantified from the 2D-FLASH lung images of mice instilled with saline or A. fumigatus spores (ASFU 1731 or FGSC A1258 strain). Error bars represent s.d. of multiple mice (saline: n=5, ASFU 1731: n=7–10, FGSC A1258: n=7–8). ***P<0.001; ****P<0.0001.
Direct comparison between the results obtained by CT and MR imaging is hampered by the dimensional discrepancy (2D vs 3D). In addition, incomplete coverage of the lungs, resulting from the inter-slice distance in the 2D-FLASH protocol, might lead to a less accurate estimation of the imaging-derived disease parameters. To further improve our self-gated imaging protocol, we optimized and evaluated a 3D-IntraGate-FLASH MRI protocol for longitudinal in vivo monitoring of IPA. Although the in-plane resolution of the acquired 3D-images was slightly lower compared with the 2D-FLASH images, the through-plane resolution was increased in combination with a full lung coverage. Hyperintense signals became clearly visible within the lungs on day 4 after infection (Figure 3a, bottom row). Scoring of these visual observations revealed that detectable, pathological changes became significant from day 3 for the FGSC A1258 strain and from day 4 for the ASFU 1731 strain (Figure 3b). These results are consistent with the results obtained by CT. The signal intensity, lung tissue volume and total lung volume increased significantly on day 4 for the FGSC A1258 strain (Figures 3c–e). The same trend was observed for the ASFU 1731 strain, although level of significance was only reached for the total lung volume on day 4. In the control group, no changes were detectable in the visual appearance of the lungs nor in the 3D-IntraGate-FLASH-derived parameters. In addition, no significant differences could be detected between the two Aspergillus strains. In accordance with the results obtained by 2D-FLASH MRI, these findings show that 3D-FLASH MRI provides both visual and quantitative information on disease development and progression in animals infected with A. fumigatus.
3D-FLASH MRI (self-gated) of the lungs after infection with Aspergillus fumigatus. (a) Representative coronal 3D-FLASH MR images of the lungs after intranasal instillation with saline (control, top row) or A. fumigatus FGSC A1258 (infection, bottom row). Dashed line depicts a manually defined region of interest (ROI) used to quantify disease progression. (b) Graph representing the cumulative image score derived from the 3D-FLASH images of mice instilled with saline or A. fumigatus spores (ASFU 1731 or FGSC A1258 strain). (c–e) Graphs representing the normalized signal intensity, lung tissue volume and total lung volume quantified from the 3D-FLASH lung images. Error bars represent s.d. of multiple mice (saline: n=3–4, ASFU 1731: n=4, FGSC A1258: n=4–8). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
UTE MRI Enables a More Sensitive Detection of Early Pathological Changes
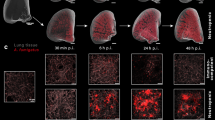
To investigate whether images acquired with an UTE would allow for a more sensitive detection of early pathological changes in the lungs, we used an optimized 3D-UTE MRI sequence to monitor the course of infection in our animals. Similar to the FLASH images, the heart and main blood vessels appeared hyperintense on the 3D-UTE MR images (Figure 4a). In contrast, signal originating from the healthy lung parenchyma could only be detected on the UTE images. Within the infected lungs, hyperintense spots became clearly discernible from the background signal originating from normal lung parenchyma on day 3 (black arrows), and increased in size and number on day 4 (Figure 4a, bottom row). Upon detailed visual scoring of the images, discrete pathological changes were already recognized on day 2 after infection, resulting in a significant increase in the cumulative score (Figure 4b). In accordance with these observations, both the signal intensity and lung tissue volume showed an increasing trend starting from day 2 after infection (Figures 4c and d). Level of significance was reached starting from day 3 for the lung tissue volume, which is 1 day earlier compared with the 2D and 3D FLASH acquisitions (Figures 2b and 3c and d) and on day 4 for the signal intensity. The total lung volume increased significantly on days 3 (FGSC A1258 strain) and 4 (both strains) after infection, which corresponds to the visual observations (Figure 4e). In addition, no significant difference could be detected between the two groups infected with the different A. fumigatus strains. These results confirm that the UTE MR sequence is more sensitive, resulting in a more efficient and earlier detection of pathological changes in the lungs of infected animals.
3D-UTE MRI (prospectively gated) of mouse lungs infected with Aspergillus fumigatus. (a) Representative coronal 3D-UTE images of the lungs after intranasal instillation with saline (control, top row) or A. fumigatus FGSC A1258 (infection, bottom row) with manually defined ROI (dashed line) indicated. Black arrows indicate hyperintense spots appearing in the infected lungs. (b) Graph representing the cumulative image score derived from the 3D-UTE images of mice instilled with saline or A. fumigatus spores (ASFU 1731 or FGSC A1258 strain). (c–e) Graphs representing the normalized signal intensity, lung tissue volume and total lung volume quantified from the 3D-UTE lung images. Error bars represent s.d. of multiple mice (saline: n=5, ASFU 1731: n=7–10, FGSC A1258: n=7–8). *P<0.05; **P<0.01; ****P<0.0001.
The In Vivo Imaging Results Are in Agreement With Gold Standard Techniques
To validate the imaging findings, clinical parameters were monitored on a daily basis. Within 4 days after infection, the animals needed to be killed for ethical reasons owing to a large drop in body weight of >25% (data not shown). In addition, the animals displayed severe clinical symptoms, such as a hunchback and reduced activity. These observations were associated with a marked increase in the imaging-derived parameters. Further validation of the imaging results was provided by the GM index in collected BAL fluid, which was significantly higher for both Aspergillus strains compared with the control group, proving that infection was successfully established (Figure 5a). The number of colonies grown from the infected lungs was significantly higher for infected animals compared with the control values, which were all zero (Figure 5b). The wet weight of lungs from animals infected with a high inoculum was significantly increased compared with the control animals, which was possibly caused by processes, such as fungal growth and edema (Supplementary Figure S1). As for the imaging results, no differences could be detected between the two groups infected with different A. fumigatus strains. Furthermore, no fungi were detected on stained lung sections of control animals (Figure 5c, right). In contrast, excessive fungal growth could be observed within the lungs of infected animals, corresponding to the large hyperintense patches seen on both CT and MR images on day 4 (Figure 5c, left). In addition, histological analysis revealed that hyphal structures mainly accumulated around the main bronchi, which corresponds to the location of lesions detected by CT and MRI, and that most airways were invaded by fungal elements (black arrows) (Figure 5d, left). Severe pulmonary injury occurred owing to invasion and destruction of the lung tissue. Large bundles of hyphae (white arrow) were observed inside pulmonary blood vessels, indicating that infection was associated with angioinvasion (Figure 5d, right). The GM index was determined on serum samples collected from a subset of animals (data not shown). All samples tested positive (OD index ≥0.5), showing the invasive character of the infection. Comparing the histological image of the left lung with the matching 3D-UTE MR images at day 4 showed clear correspondence between affected areas identified by both techniques (Figure 5e). For example, when the entire proximal part of the lungs appeared hyperintense on the 3D-UTE MR images, the corresponding regions turned out to be completely infected on histology (Figure 5e, left). On the other hand, if hyperintense signals were centered around the main bronchi on the MR images, most fungal foci were also centered around the main bronchi on histology (Figure 5e, right). However, the affected areas can more easily be distinguished on histology by a non-trained person, while radiological skills are needed for the correct interpretation of the MR and CT images at the onset of infection. Likewise, the affected areas identified by histology corresponded well with hyperintense areas detected on the 3D-FLASH MR and CT images (Figures 1a and 3a). These results confirm the presence of infection in all animals and support the results obtained by both imaging modalities.
Validation of the imaging results. (a, b) Graphs representing individual galactomannan levels in BAL fluid and the individual fungal loads on day 4 after infection (saline: n=5, ASFU 1731: n=6–7, FGSC A1258: n=5–7). (c) Light microscopic image of a representative PAS-stained lung section 4 days after instillation with FGSC A1258 (Infection (5 × 105 spores), left) or saline (Control, right). Scale bar (upper left corner) measures 50 μm. (d) Light microscopic image of PAS-stained lung section showing fungal hyphae lining the main airways (left image, black arrows) and invading a pulmonary blood vessel (right image, white arrow). Scale bar measures 200 μm. (e) Representative coronal UTE MR images of the lungs 4 days after infection with FGSC A1258. Left hand side (L) and right hand side (R) are indicated on the image. Histological images of the corresponding left lung lobe are next to the corresponding MR image. Scale bars measure 2 mm. ***P<0.001.
Imaging Allows for Sensitive Detection of Differences in Disease Development Between Individual Animals Upon Infection with a Small Inoculum
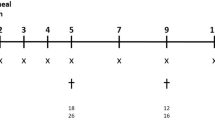
To investigate whether non-invasive imaging techniques can be used to detect differences in disease development between individual animals challenged with a smaller inoculum, mice were infected with an inoculum of 2 × 105 conidia (FGSC A1258 strain) and monitored daily with CT and MR imaging (3D-UTE and 3D-FLASH). Evaluation of clinical parameters showed that humane end points were reached either on day 4 (n=1), day 5 (n=4), day 6 (n=1) or day 7 (n=1) after infection. Visual assessment of the MR and CT images demonstrated the presence of pathological changes within the lungs of all animals, resulting in a gradual increase of the cumulative image score (Figures 6a–c(A)). In addition, the total lung volume showed an increasing trend for almost all animals on the MR and CT images, reaching maximum values on day 5 or 6 after infection (Figures 6a–c(D)). As expected, these results show that the infection develops more slowly with a lower inoculum compared with the animals with a higher inoculum. Furthermore, quantification of the signal intensity and lung tissue volume showed a remarkable discrepancy between groups of animals infected with a low inoculum. For approximately half of all animals (subgroup A), a large increase in signal intensity and lung tissue volume was observed on the last day of follow-up (Figures 6a–c(B and C)). For the other half of the animals (subgroup B), these parameters remained stable over time. Thus imaging revealed that not all animals infected with the small inoculum developed IPA to the same extent as that of the animals infected with the large inoculum. Histological analysis showed that the lungs of animals in subgroup A were extensively damaged by fungal invasion consistent with the levels of their imaging-derived parameters, while damage in the lungs of the other animals (subgroup B) was more restricted (Figure 6d). In addition, the GM index and the number of viable colonies was significantly higher for the animals in subgroup A compared with the animals in subgroup B, supporting the imaging findings (Figures 6e and f). Similar results were obtained by using the ASFU 1731 strain (data not shown). Solely based on the imaging results, we could discriminate between differences in disease severity for individual animals.
CT and MR lung imaging of mice infected with a small inoculum (2 × 105 conidia; FGSC A1258 strain). Graphs representing the cumulative image score (A), normalized signal intensity (B), lung tissue volume (C) and total lung volume (D) quantified for individual animals from the 3D-UTE MR images (a), 3D-FLASH MR images (b) and CT images (c). The animals that developed more severe infection were assigned to subgroup A; the animals that developed less severe infection were assigned to subgroup B. Note that the last CT data point of two animals from subgroup A and two animals from subgroup B are missing as these animals were only scanned by MRI at the last time point owing to acute worsening of their general state following anesthesia. (d) Light microscopic image of a PAS-stained lung section from subgroup A (left) and subgroup B (right). Scale bar measures 2 mm. (e, f) Graphs representing individual galactomannan levels in BAL fluid (1/1000 dilution) and fungal load. Error bars represent s.d. of subgroup A (n=3) or subgroup B (n=4). *P<0.05; ***P<0.001.
DISCUSSION
In this study, it was shown that non-invasive CT and MR imaging provides both visual information and multiple quantitative parameters describing disease progression in mice suffering from IPA. Pathogen-related changes were clearly detectable within infected lungs on both CT and MR images. These pathological changes were visualized as extensive, homogeneous lesions that were mainly localized in the proximal part of the lungs and centered around the main airways. Our visual observations were supported by a significant increase in (semi)quantitative image scores, showing that CT and MR imaging are equally sensitive in detecting extensive lesions present in an advanced stage of infection. The use of advanced MRI (3D-UTE) enabled an earlier, more sensitive detection of lesions developing within the lungs compared with more conventional gradient echo (FLASH) MR and CT imaging. These results support findings from previous studies that also reported on the increased sensitivity of UTE MRI for the identification of lung lesions.36, 37 Such techniques with an enhanced sensitivity provide valuable information on both early and late stages of infection on an individual level, which is of great importance when using an animal model that is characterized by a high variability and a very short time window between disease onset and death. In addition, UTE MRI can be very useful in preclinical drug screening studies, for example, to identify the optimal time point to begin with an antifungal treatment based on individual data.
Visual scoring of the CT and MR images proved to be a reliable strategy for obtaining rapid, semiquantitative information on disease progression. In previous studies, a similar scoring system was used to obtain semiquantitative information on the development of pulmonary lesions based on ultrafast CT lung images of rabbits suffering from IPA.40, 41, 42 However, radiological skills are mandatory and the method is to some extent observer-dependent. To obtain an objective read-out for the developing infection, multiple lung parameters were quantified based on a VOI covering the entire lung. The lung tissue volume and signal intensity, two parameters that are directly associated with lesion development, showed a gradual increase over time in both imaging modalities, associated with progressing infection. Again, the high sensitivity of UTE MRI for detecting subtle pathological changes was reflected in an early increase in both measures, whereas the sensitivity of FLASH MRI to quantitatively detect pulmonary lesions proved to be lower. Compared with FLASH MRI, CT imaging proved to be slightly more sensitive for the quantitative detection of pulmonary lesions, providing excellent air–tissue contrast and high resolution. Nevertheless, CT proved to be less sensitive for the early detection of pathological changes within the lungs compared with UTE MRI. The superiority of UTE MRI in providing visual and quantitative information on disease progression is hereby confirmed.
Upon both visual and quantitative assessment of the CT and MR images, an increase in the size of the lungs as a whole was revealed. A similar increase in total lung volume was also observed in other lung disease models, such as the bleomycin-induced lung fibrosis model43 and pulmonary cryptococcosis model. This indicates that total volume changes are not specifically related to the development of a pulmonary fungal infection. Based on the observation that the total lung volume increases later in time compared with the other changes, it could be considered as a consequence rather than a direct indicator of progressing lung disease. However, further studies are needed to gain more insight into the exact role of the observed total lung volume changes in lung disease models.
Upon infection with a small inoculum, an increased variability in disease development was observed between individual mice. Although most animals displayed similar symptoms, longitudinal CT and MR imaging revealed that one half of the animals developed extensive lesions within the lungs, accompanied by a marked increase in all quantified lung parameters in the advanced stages of the disease. On the other hand, reduced lesion burden was detected within the lungs of the other half of animals, associated with unaltered signal intensities and lung tissue volumes over time. Solely based on these imaging findings, animals could be divided into two subgroups corresponding to animals that developed IPA and animals that were able to better cope with the pathogens. Validation of the imaging results showed a clear difference between these two groups, thereby supporting the imaging findings. These inter-individual differences, which are possibly related to a heterogeneous spore disposition upon intranasal infection with a low inoculum, are of high importance for the correct interpretation of results, especially in studies evaluating the effect of antifungal drugs. If disease develops more slowly in a subset of animals, the presence of smaller lesions after therapy might be falsely interpreted as a positive effect from the antifungal treatment, even if this was not the case. In addition, if no lesions were present before the start of the therapy owing to successful clearance of the infection by the host immune system, the lack of disease could again be falsely attributed to an assumed effect of the antifungal treatment. Therefore, disease should be confirmed before treatment is initiated to ensure that the observed improvements are directly related to the administered treatment and extent of initial disease. However, animals that do not develop extensive lesions within the lungs should not be excluded from treatment studies, as they can provide better insight into the variations in responses to antifungal therapy. Thus, by using non-invasive imaging techniques such as CT or MRI, the infectious state of the animal can be assessed repeatedly over time and any changes in lesion volume can be compared with reference images of the same animal acquired at earlier time points. In addition, non-invasive imaging can be used to determine an objective time point for starting treatment, thereby enabling the differential evaluation of prophylactic vs truly therapeutic treatment regimes. This approach will be most effective if lesions are detected at an early stage of infection, as there is a higher possibility that antifungal therapy will be successful upon early initiation. However, as this approach would imply that individual animals within one experimental group would receive a certain treatment at different time points, one must be careful with interpreting the results. This potential challenge could be overcome by using sufficiently large experimental groups, so that a statistically relevant comparison of drug efficacy between these various subgroups can be achieved. Nevertheless, our observations clearly demonstrate that non-invasive imaging techniques could have a crucial role in improving preclinical in vivo drug screening processes.
All imaging results were validated based on three methods currently regarded as gold standards in clinical microbiology. The presence of fungi in the lungs was confirmed by all techniques. As expected, it proved to be very difficult to obtain reliable quantitative information on disease severity based on CFU counting. Large inter-animal variability was observed in the infection groups, which did not correspond to the variation detected by imaging and histology. Most likely, this variation resulted from difficulties in growing colonies from the lung tissue owing to hyphal growth. In two cases, not even one single colony could be grown from the tissue despite positive GM and histology results, underlining the limitations of this technique. In contrast, CT and MRI provided reliable quantitative readouts reflecting the infectious state of the animals at different time points. Based on histology, the invasive character of IPA was proven by bundles of hyphae penetrating the airways and blood vessels. This was also confirmed by GM antigen detection in serum. To assess the agreement between histology and imaging, histological sections at a certain position within the lung were visually compared with the corresponding MR and CT images. The distribution of fungal lesions corresponded very well between histology and imaging, indicating that imaging provides reliable information on disease-related changes within the lungs. However, solely based on the acquired CT and MR images, it proved to be difficult to distinguish fungal mass from edema and/or necroses within the lungs. Further studies are needed to investigate the feasibility of combining CT or MRI with optical imaging techniques to obtain additional information on different disease processes within the lungs. For now, it remains important to validate the results obtained by these non-invasive imaging techniques with standard techniques, such as histology.
By introducing non-invasive imaging in preclinical studies, dynamic information on disease development can be obtained without the need to sacrifice numerous animals at every time point. As a result, the required number of animals and the associated costs can drastically be reduced, provided that one has access to the equipment and expertise needed to acquire and interpret the imaging data. One has to be cautious with the interpretation of the imaging data as the imaging-derived parameters are more phenotypic markers, not necessarily specific to aspergillosis. Therefore, they should not be mistaken as disease specific, potentially diagnostic biomarkers. In addition, future studies need to be carried out to determine whether potential differences in pathogenesis between different Aspergillus strains can be investigated efficiently by using a non-invasive imaging approach.
In conclusion, we show that CT and MR imaging techniques are highly valuable for the longitudinal assessment of dynamic infectious disease-related changes in individual animals, in contrast to standard invasive techniques that only provide a snapshot image of the infection. Non-invasive imaging provides crucial insight in inter-individual differences in IPA development, which can have a major influence on preclinical drug screening processes. Testing novel antifungal drugs may become more efficient, more ethical and possibly less expensive by introducing non-invasive imaging in these kinds of studies.
References
Walsh TJ, Anaissie EJ, Denning DW et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis 2008;46:327–360.
Pasqualotto AC . Aspergillosis: From Diagnosis to Prevention. Springer: Dordrecht, The Netherlands, 2010.
Latge JP . Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 1999;12:310–350.
Goodley JM, Clayton YM, Hay RJ . Environmental sampling for aspergilli during building construction on a hospital site. J Hosp Infect 1994;26:27–35.
Dagenais TR, Keller NP . Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 2009;22:447–465.
Bitar D, Lortholary O, Le Strat Y et al. Population-based analysis of invasive fungal infections, France, 2001-2010. Emerg Infect Dis 2014;20:1149–1155.
Pagano L, Caira M, Candoni A et al. Invasive aspergillosis in patients with acute myeloid leukemia: a SEIFEM-2008 registry study. Haematologica 2010;95:644–650.
Baddley JW, Andes DR, Marr KA et al. Factors associated with mortality in transplant patients with invasive aspergillosis. Clin Infect Dis 2010;50:1559–1567.
Snelders E, van der Lee HA, Kuijpers J et al. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 2008;5:e219.
Howard SJ, Cerar D, Anderson MJ et al. Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 2009;15:1068–1076.
Bueid A, Howard SJ, Moore CB et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother 2010;65:2116–2118.
van der Linden JW, Arendrup MC, Warris A et al. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg Infect Dis 2015;21:1041–1044.
Vermeulen E, Maertens J, De Bel A et al. Nationwide surveillance of azole resistance in Aspergillus disease. Antimicrob Agents Chemother 2015;59:4569–4576.
Takemoto K, Yamamoto Y, Ueda Y et al. Comparative studies on the efficacy of AmBisome and Fungizone in a mouse model of disseminated aspergillosis. J Antimicrob Chemother 2004;53:311–317.
Olson JA, Schwartz JA, Hahka D et al. Toxicity and efficacy differences between liposomal amphotericin B formulations in uninfected and Aspergillus fumigatus infected mice. Med Mycol 2015;53:107–118.
Brock M, Jouvion G, Droin-Bergere S et al. Bioluminescent Aspergillus fumigatus, a new tool for drug efficiency testing and in vivo monitoring of invasive aspergillosis. Appl Environ Microbiol 2008;74:7023–7035.
Morisse H, Heyman L, Salaun M et al. In vivo and in situ imaging of experimental invasive pulmonary aspergillosis using fibered confocal fluorescence microscopy. Med Mycol 2012;50:386–395.
Galiger C, Brock M, Jouvion G et al. Assessment of efficacy of antifungals against Aspergillus fumigatus: value of real-time bioluminescence imaging. Antimicrob Agents Chemother 2013;57:3046–3059.
Vande Velde G, Kucharikova S, Schrevens S et al. Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cell Microbiol 2014;16:115–130.
Brock M . Application of bioluminescence imaging for in vivo monitoring of fungal infections. Int J Microbiol 2012;2012:956794.
Petrik M, Franssen GM, Haas H et al. Preclinical evaluation of two 68Ga-siderophores as potential radiopharmaceuticals for Aspergillus fumigatus infection imaging. Eur J Nucl Med Mol Imaging 2012;39:1175–1183.
Wang Y, Chen L, Liu X et al. Detection of Aspergillus fumigatus pulmonary fungal infections in mice with (99 m)Tc-labeled MORF oligomers targeting ribosomal RNA. Nucl Med Biol 2013;40:89–96.
Severin GW, Jorgensen JT, Wiehr S et al. The impact of weakly bound Zr on preclinical studies: non-specific accumulation in solid tumors and aspergillus infection. Nucl Med Biol 2014;42:360–368.
Suetens P . Fundamentals of Medical Imaging. Cambridge University Press: New York, USA, 2009.
Clark DP, Badea CT . Micro-CT of rodents: state-of-the-art and future perspectives. Phys Med 2014;30:619–634.
De Langhe E, Vande Velde G, Hostens J et al. Quantification of lung fibrosis and emphysema in mice using automated micro-computed tomography. PLoS One 2012;7:e43123.
Rodt T, von Falck C, Dettmer S et al. Micro-computed tomography of pulmonary fibrosis in mice induced by adenoviral gene transfer of biologically active transforming growth factor-beta1. Respir Res 2010;11:181.
Li M, Jirapatnakul A, Biancardi A et al. Growth pattern analysis of murine lung neoplasms by advanced semi-automated quantification of micro-CT images. PLoS One 2013;8:e83806.
Artaechevarria X, Blanco D, de Biurrun G et al. Evaluation of micro-CT for emphysema assessment in mice: comparison with non-radiological techniques. Eur Radiol 2011;21:954–962.
Fushiki H, Kanoh-Azuma T, Katoh M et al. Quantification of mouse pulmonary cancer models by microcomputed tomography imaging. Cancer Sci 2009;100:1544–1549.
McRobbie D, Moore E, Graves M et al. MRI from Picture to Proton. Cambridge University Press: Cambridge, UK, 2003.
Beckmann N, Tigani B, Mazzoni L et al. Techniques: magnetic resonance imaging of the lung provides potential for non-invasive preclinical evaluation of drugs. Trends Pharmacol Sci 2003;24:550–554.
Babin AL, Cannet C, Gerard C et al. Noninvasive assessment of bleomycin-induced lung injury and the effects of short-term glucocorticosteroid treatment in rats using MRI. J Magn Reson Imaging 2011;33:603–614.
Babin AL, Cannet C, Gerard C et al. Bleomycin-induced lung injury in mice investigated by MRI: model assessment for target analysis. Magn Reson Med 2012;67:499–509.
Wang F, Akashi K, Murakami Y et al. Detection of lung tumors in mice using a 1-Tesla compact magnetic resonance imaging system. PLoS One 2014;9:e94945.
Bianchi A, Dufort S, Fortin PY et al. In vivo MRI for effective non-invasive detection and follow-up of an orthotopic mouse model of lung cancer. NMR Biomed 2014;27:971–979.
Vande Velde G, De Langhe E, Poelmans J et al. Magnetic resonance imaging for noninvasive assessment of lung fibrosis onset and progression: cross-validation and comparison of different magnetic resonance imaging protocols with micro-computed tomography and histology in the bleomycin-induced mouse model. Invest Radiol 2014;49:691–698.
Takahashi M, Togao O, Obara M et al. Ultra-short echo time (UTE) MR imaging of the lung: comparison between normal and emphysematous lungs in mutant mice. J Magn Reson Imaging 2010;32:326–333.
Vande Velde G, De Langhe E, Poelmans J et al. Longitudinal in vivo micro-computed tomography of mouse lungs: no evidence for radiotoxicity. Am J Physiol Lung Cell Mol Physiol 2015;309:L271–L279.
Petraitis V, Petraitiene R, Sarafandi AA et al. Combination therapy in treatment of experimental pulmonary aspergillosis: synergistic interaction between an antifungal triazole and an echinocandin. J Infect Dis 2003;187:1834–1843.
Walsh TJ, Petraitis V, Petraitiene R et al. Diagnostic imaging of experimental invasive pulmonary aspergillosis. Med Mycol 2009;47 (Suppl 1):S138–S145.
Petraitiene R, Petraitis V, Groll AH et al. Antifungal efficacy of caspofungin (MK-0991) in experimental pulmonary aspergillosis in persistently neutropenic rabbits: pharmacokinetics, drug disposition, and relationship to galactomannan antigenemia. Antimicrob Agents Chemother 2002;46:12–23.
Egger C, Gerard C, Vidotto N et al. Lung volume quantified by MRI reflects extracellular-matrix deposition and altered pulmonary function in bleomycin models of fibrosis: effects of SOM230. Am J Physiol Lung Cell Mol Physiol 2014;306:L1064–L1077.
Acknowledgements
This work was supported by funding provided by the Flemish research foundation (FWO; 15061.14N, G.0691.15N and G.0804.11) and KU Leuven ((PF 10/017 (IMIR)) and BOF-CREA/14/015). JP received a PhD grant for strategic basic research from the Agency for Innovation by Science and Technology (IWT). GVV received a postdoctoral fellowship from the FWO. The FGSC A1258 A. fumigatus strain was obtained from the fungal genetics stock center (Kansas City, MO, USA). We thank Professor Enrico Radaelli for optimizing the histology protocol and Lorna Omodho for her excellent technical assistance (Histology Unit, Center for Human Genetics and VIB11, Center for the Biology of Disease, KU Leuven, Belgium).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
To enable non-invasive, longitudinal monitoring of pulmonary aspergillosis, the authors optimized computed tomography and magnetic resonance imaging techniques with A. fumigatus-infected mice. These techniques may prove to be valuable tools for the evaluation of dynamic disease-related changes and differences in disease severity in individual animals, and might be applied as rapid and cost-efficient drug screening protocols in preclinical models.
Supplementary information
Rights and permissions
About this article
Cite this article
Poelmans, J., Hillen, A., Vanherp, L. et al. Longitudinal, in vivo assessment of invasive pulmonary aspergillosis in mice by computed tomography and magnetic resonance imaging. Lab Invest 96, 692–704 (2016). https://doi.org/10.1038/labinvest.2016.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.45
This article is cited by
-
Longitudinal micro-computed tomography-derived biomarkers quantify non-resolving lung fibrosis in a silicosis mouse model
Scientific Reports (2020)
-
Differentiating lung cancer and infection based on measurements of extracellular pH with acidoCEST MRI
Scientific Reports (2019)
-
Bronchoscopic fibered confocal fluorescence microscopy for longitudinal in vivo assessment of pulmonary fungal infections in free-breathing mice
Scientific Reports (2018)