Abstract
The miR-19 family (miR-19a and miR-19b-1) are key oncogenic components of the miR-17-92 cluster. Overexpression of miR-19 is strongly associated with cancer invasion and metastasis, and poor prognosis of cancer patients. However, the underlying mechanisms remain largely unknown. In the present study, we found that enforced expression of miR-19 including miR-19a and miR-19b-1 triggered epithelial–mesenchymal transition (EMT) of lung cancer cells A549 and HCC827 as shown by mesenchymal-like morphological conversion, downregulation of epithelial proteins (e.g., E-cadherin, ZO-1 (zona occludens 1), and α-catenin), upregulation of mesenchymal proteins (e.g., vimentin, fibronectin 1, N-cadherin, and snail1), formation of stress fibers, and reduced cell adhesion. In addition, enhanced migration and invasion were observed in the cancer cells A549 and HCC827 undergoing EMT. In contrast, silencing of endogenous miR-19 reversed EMT and reduced the migration and invasion abilities of A549 and HCC827 cells. DNA microarray results revealed significant changes of the expression of genes related to EMT, migration, and metastasis of miR-19-expressing A549 cells. Moreover, siRNA-mediated knockdown of PTEN, a target of miR-19, also resulted in EMT, migration, and invasion of A549 and HCC827 cells, suggesting that PTEN is involved in miR-19-induced EMT, migration and invasion of lung cancer cells. Furthermore, lung cancer cells undergoing EMT induced by miR-19 demonstrated reduced proliferation in vitro and in vivo, and enhanced resistance to apoptosis caused by TNF-α. Taken together, these findings suggest that miR-19 triggers EMT, which has an important role in the invasion and migration of lung cancer cells, accompanied by the reduced proliferation of cells.
Similar content being viewed by others
Main
It has been reported that microRNAs (miRNAs) have critically important roles in carcinogenesis, cancer invasion and metastasis, and tumor angiogenesis.1, 2, 3, 4 The miR-17-92 gene cluster encodes six miRNAs of four miRNA families: the miR-17 family including miR-17-5p and miR-20a, the miR-18 family (miR-18a), the miR-19 family (miR-19a and miR-19b-1), and the miR-92 family. Upregulation of these miRNAs have been widely reported in B-cell lymphoma and numerous solid tumors, such as lung, breast, colon, pancreatic, prostate, and stomach cancers.1, 5 For e.g., the interaction between miR-17-92 and c-Myc contributed to the development of lymphomas in mice.1 In addition, enforced expression of the miR-17-92 gene cluster promoted tumor cell proliferation of lung cancer,6 esophageal cancer,7 and hepatocellular carcinoma,8 suggesting the oncogenic role of miR-17-92. Among the mir-17-92 gene cluster, the miR-19 family were considered as key oncogenic components based on experiments in mouse models of lymphoma.9, 10 The present study aimed to investigate the role and mechanism of miR-19 in the progression of lung cancer.
Previous studies have also reported that upregulation of miR-19a and miR-19b-1 was associated with cancer invasion and metastasis, and poor prognosis of patients with early non-small-cell lung carcinoma (NSCLC),11, 12, 13 gastric cancer,14 and esophageal squamous cell carcinoma.15 In addition, high levels of serum miR-19a and miR-19b-1 were considered as independent prognostic factors for poor survival of NSCLC patients.12, 13 In a mouse model of c-Myc-driven B-cell lymphomas, Em-myc/miR-19b-1 lymphomas are highly invasive as lymphoma cells invaded the thymus and bone marrow, as well as visceral organs outside the lymphoid compartment, including the liver, lung, and, occasionally, the kidney.10 More recent studies have reported that miR-19a or miR-19b-1 promoted the migration and invasion of cervical carcinoma cells,16 colon cancer cells,17 and gastric cancer cells through targeting the tumor suppressor MXD1,14 and Hela, MCF7, and Huh7 cells through targeting TP53.18 Taken together, these results support that miR-19 is involved in cancer invasion and metastasis; however, the underlying mechanisms remain largely unknown.
Interestingly, we observed in a preliminary experiment that lung cancer cells with miR-19 (miR-19a/miR-19b-1) overexpression exhibited typical spindle-shaped and fibroblast-like morphology, suggesting the occurrence of epithelial–mesenchymal transition (EMT).
EMT is a central mechanism contributing to the invasion and metastasis of various cancers.19, 20 Therefore, identification of key factors involved in EMT and investigation of the molecular mechanisms underlying EMT are of critical importance for understanding tumor invasion and metastasis, and developing novel interventions for the treatment of metastatic cancers.
Given that miR-19 is associated with cancer invasion and metastasis and EMT is the central mechanism underlying cancer invasion and metastasis,10, 11, 12, 13, 14, 15, 16, 17, 18 the present study aimed to investigate whether miR-19 is able to induce EMT and enhance invasion and metastasis of human cancer cells. Our results showed that enforced expression of miR-19 in lung cancer cells induced EMT, which caused growth inhibition but elevated migration of the cancer cells.
MATERIALS AND METHODS
Establishment of Lung Cancer Cell Lines Stably Expressing miR-19
The human lung cancer cell line A549-luc with luciferase (Luc) expression was purchased from Xenogen Corporation (MA, USA). The human lung cancer cell line HCC827 was obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The A549 and HCC827 cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS). Construction of the lentiviral vector of pLV-miR-19 and the recombinant lentiviruses LV-con and LV-miR-19 were conducted according to the protocol described in the Supplementary Methods. The recombinant lentiviruses were then used to infect A549 and HCC827 cells. Successful overexpression of miR-19 was verified by quantitative real-time RT-PCR (qRT-PCR) (data not shown).
miRNA Transient Transfection
The human miR-19 inhibitor (miR-19a inhibitor and miR-19b-1 inhibitor) and a nonspecific miRNA inhibitor control (i.e., inhibitor control) were all purchased from Shanghai GenePharma (Shanghai, China). MiRNAs were transiently transfected into cells at a working concentration of 100 nmol/l using lipofectamine 2000 reagent (Invitrogen, CA, USA) in accordance with the manufacturer’s procedure. Cells for total RNA isolation and the transwell migration assay or Boyden invasion assay were harvested 48 h after oligonucleotide treatment.
RNA Isolation and qRT-PCR
Total RNA was extracted from lung cancer cells using the Trizol reagent (TaKaRa) and reversely transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa) according to the manufacturer’s instruction. To quantify the expression of miR-19 and protein-coding genes, qRT-PCR was performed using a SYBR Green qRT-PCR master mix kit (TaKaRa) on a Stratagene Mx3000P qRT-PCR System according to the manufacturer’s instruction. The primers used in qRT-PCR assay were listed in Supplementary Tables S1–S3. To measure the level of miR-19, U6 was used as an internal control, whereas GAPDH was used as an internal control for the mRNA expression analysis of protein-coding genes. All samples were normalized based on internal controls, and fold changes were calculated through relative quantification (2−ΔΔCt).
Western Blot Analysis
Total proteins were extracted from A549 and HCC827 cells, and western blot was conducted as previously described. Briefly, extracted proteins were separated by SDS-PAGE, and electrophoretically transferred to a polyvinylidene difluoride membrane. The blots were probed with primary antibodies and then horseradish peroxidase-labeled goat-antimouse IgG. Hybridization signal was detected using enhanced chemiluminescence. To measure the expression of different proteins, β-actin was used as a loading control. The antibodies used in this study are shown in Supplementary Table S4.
Transwell Migration Assay and Boyden Invasion Assay
For the transwell migration assay, tumor cells (1 × 105) were seeded into the upper chamber (BD Biosciences, MA) with serum-free DMEM. Boyden invasion assay was conducted with matrigel (BD Biosciences) in the upper chamber. DMEM with 10% FBS was added into lower compartment as chemoattractant. A549 cells were allowed to migrate for 14 and 24 h in transwell migration assay and Boyden invasion assay, respectively. HCC827 cells were allowed to migrate for 22 and 46 h in transwell migration assay and Boyden invasion assay, respectively. Remaining cells in the upper chamber were removed using a cotton swap. The cells migrated to the lower surface of the membrane were fixed with 100% methanol, stained with hematoxylin, and counted in five randomly selected optical fields under a microscope.
Cell Adhesion Assay
Cell adhesion assay was performed according to the protocol previously described.21 Briefly, miR-19- and vector-expressing A549 cells were seeded onto culture dishes and grown to ~90% confluence. The confluent monolayers were then washed with PBS and treated with 0.05 mM EDTA. Cells detachment was examined at different time intervals (2, 20, 30, and 40 min) under a phase contrast microscope and cell adhesion was evaluated by counting the adherent cells.
Microarray Analysis
Microarray analysis was carried out using the Affymetrix Human Gene U133 Plus 2.0 array according to the manufacturer’s standard protocol. This array contains 47 000 transcripts from 38 500 well-characterized human genes. RNA preparation was conducted according to the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, total RNA was extracted from frozen samples using TRIZOL reagents, purified, and concentrated using the RNeasy MinElute Cleanup kit and an on-column DNase treatment, respectively. RNA was quantified and qualified using a UV spectrophotometer at 260 and 280 nm. RNA integrity was checked by electrophoresis (1% formaldehyde denaturing gel) and stained with ethidium bromide. DNA labeling, hybridization, washing, and scanning were performed according to the standard operating procedure of CapitalBio Corporation. Briefly, total RNA was used to synthesize cDNA that was fluorescently labeled with Cy5 or Cy3-CPT using Klenow enzyme. Labeled cDNA was hybridized with probes on the Affymetrix Human Gene U133 Plus 2.0 array at 45 °C for 16 h in an Affymetrix GeneChip Hybridization Oven 640. Array slides were then washed and stained in the Affymetrix Fluidics Station 450. Subsequently, array slides were scanned using Affymetrix GeneChip Scanner 3000, and the obtained images were analyzed using the LuxScan Version 3.0 (CapitalBio Corporation). LOWESS method was used for the normalization. A total of three biological replicates were performed.
Statistical t-test and multiple test corrections were conducted to identify genes with significant expression (increase or decrease). Significantly expressed genes were defined as genes with a false discovery rate <0.01 and twofold or higher change of expression based on three independent biological replicates. Data analysis was performed using the Significance Analysis of Microarray software (SAM 3.0, Stanford University, USA; http://www-stat.stanford.edu). Significantly expressed genes were grouped into functional categories according to the Gene Ontology (GO) information and analyzed using the Molecule Annotation System (http://bioinfo.capitalbio.com/mas3/). Biological pathways were identified according to known pathways in the KEGG database (http://www.genome.jp/kegg/). To validate the microarray results, qRT-PCR was performed to evaluate the expression of a number of significantly expressed genes identified through microarray analysis.
Luciferase Reporter Assay
The dual luciferase reporter gene plasmid (i.e., pLuc-PTEN-3′-UTR-wt) containing the putative miR-19-binding site at the 3′-UTR of PTEN mRNA was purchased from Kangbio (Shenzhen, China). Cells were seeded in 48-well plates and cultured for 48 h. The pLuc-PTEN-3′-UTR-wt plasmid was co-transfected into A549 with the miR-19a mimics, mimics control, miR-19a inhibitor, or inhibitor control using lipofectamine 2000 reagent (Invitrogen), respectively. Luciferase and Renilla activities were evaluated 48 h post transfection using the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions. All experiments were performed independently in triplicate.
RNA Interference (RNAi)
RNAi was used to knock down PTEN expression. The specific siRNA duplexes targeting the human PTEN gene (sense: 5′-GAGCGUGCAGAUAAUGACAdTdA-3′; antisense: 3′-dAdTCUCGCACGUCUAUUACUGU-5′) were synthesized by RiboBio (Guangzhou, China). The siRNA duplexes with nonspecific sequence were used as a negative controls. SiRNA transfection was carried out with lipofectamine 2000 (Invitrogen) according to the manufacture’s instruction. Transfection efficiency was evaluated by western blot.
MTT Assay and Cell-Cycle Analysis
A549 cells infected with either LV-con or LV-miR-19 were plated in 96-well plates (1 × 103 cells/well) in a final volume of 200 μl, and then cultured for 1, 2, 3, 4, 5, 6, and 7 days. MTT assay was used to evaluate the effects of miR-19 on cell growth. For cell-cycle analysis, A549 cells infected with either LV-con or LV-miR-19 were plated in six-well plates (2 × 105 cells/well). At 48 h post plating, cells were stained with propidium iodide (PI) and the cell cycle was analyzed using flow cytometry.
Colony Formation Assay
A549 and HCC827 cells infected with either LV-con or LV-miR-19 were counted, plated at 200 cells for the pooled population in six-well plates, and cultured with RPMI1640 complete culture medium for 10 days. Colonies were fixed with methanol and stained with hematoxylin. The plates were photographed after washing off the dye and the colony was counted under the microscope. At least three independent experiments were carried out for each assay.
Animal Tumor Models
The animal experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Southern Medical University. The animal protocol was approved by the Committee on Ethics of Animal Experiments of the Southern Medical University (approval number: 2013-0012). Female BALB/c nude mice (4–5 weeks) were purchased from the Center of Experimental Animals, Southern Medical University, and housed in microisolator cages under aseptic conditions. A549 cells that stably express miR-19 (5 × 106 cells) or vector control (5 × 106 cells) were subcutaneously injected into the right or left dorsal thigh of mice (n=6). All surgeries were performed under sodium pentobarbital anesthesia, and all efforts were made to minimize the suffering of animals.
Analysis of Caspase-3 and Caspase-8 Activities
The activities of caspase-3 and caspase-8 were evaluated using the commercial caspase-3 and -8 activity assay kits (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer’s instruction. Cells attached to the dish and in the supernatant were collected and lysed in 5 mM Tris-HCl (pH 8.0), 20 mM EDTA, and 0.5% Triton X-100 at 4 °C. Ac-DEVD-pNA and Ac-IETD-pNA were used as substrates to measure the enzymatic activities of caspase-3 and caspase-8, respectively. Enzymatic activity assays were conducted on 96-well microtitre plates by incubating 10 μl proteins from cell lysate per sample in 80 μl reaction buffer (1% NP-40, 20 mM Tris-HCl (pH 7.5), 137 mM Nad, and 10% glycerol) containing 10 μl caspase-3 substrate (Ac-DEVD-pNA, 2 mM) or caspase-8 substrate (Ac-IETD-pNA, 2 mM). Lysates were incubated at 37 °C for 4 h. Samples were measured on an ELISA reader (Bio-Tek ELX800) at the absorbance of 405 nm. The protein concentration of cell lysates was determined using the Bradford protein assay kit, and the results were presented as units of caspase activity per μg of protein.
Statistical Analysis
Data were presented as mean±s.d. unless otherwise indicated of at least three independent experiments. Statistical analysis was performed using a SPSS 13.0 software package. Two-tailed Student’s t-test was used for comparisons between two independent groups. Statistical significance was assessed by the Student’s t-test (#P<0.05; *P<0.01).
RESULTS
miR-19 Overexpression Induced EMT-Like Cellular Marker Alterations in A549 and HCC827 Cells
We first evaluated the expression of miR-19a and miR-19b-1 in available nine lung cancer cell lines (i.e., H358, H1650, A549, Spc-A-1, HCC827, 95D, H23, H446, and H460 cells) and the BEAS-2B cell line (a normal human bronchial epithelial cell line) based on qRT-PCR. The results showed that the expression of miR-19a and miR-19b-1 was significantly higher expressed in lung cancer cell lines than that in BEAS-2B cells (Supplementary Figure S1).
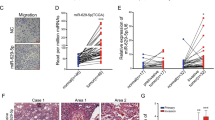
Then, miR-19a and miR-19b-1 were ectopically expressed in A549 and HCC827 cells to investigate the biological function of the miR-19 family in the development of lung cancer. Surprisingly, ectopic expression of miR-19 in A549 and HCC827 cells led to the occurrence of spindle-like and fibroblastic morphology (Figure 1a), suggesting that miR-19-expressing A549 and HCC827 cells might have undergone EMT. To confirm that miR-19 induced EMT, we examined the expression of a number of epithelial and mesenchymal markers. First, qRT-PCR results showed that miR-19 expression significantly downregulated the epithelial marker E-cadherin and upregulated the mesenchymal markers vimentin, fibronectin 1 (FN1), N-cadherin, and snail1 in both A549 and HCC827 cells (Figure 1b). In addition, western blot analysis also confirmed the reduced expression of E-cadherin, ZO-1, and α-catenin, as well as increased expression of mesenchymal markers (i.e., vimentin, FN1, N-cadherin, and snail1) in miR-19-expressing A549 and HCC827 cells (Figure 1c). Moreover, immunohistochemistry staining and western blot analysis revealed significantly higher expression of FN1, N-cadherin, and vimentin, and significantly lower expression of E-cadherin in the xenografts of miR-19-expressing A549 cells in nude mice, compared with xenografts derived from vector-expressing A549 cells (Supplementary Figure S9). To further examine the effects of loss of miR-19 function on EMT of lung cancer cells, endogenous miR-19a and miR-19b-1 in A549 and HCC827 cells were silenced using miR-19a inhibitor and miR-19b-1 inhibitor (Supplementary Figure S2), respectively. Downregulation of endogenous miR-19a and miR-19b-1 in A549 and HCC827 cells, respectively, increased the expression of epithelial marker (i.e., E-cadherin) and concomitantly reduced the expression of mesenchymal markers (i.e., vimentin, FN1, and N-cadherin) (Supplementary Figure S2). Given that activation of the PI3K/Akt pathway is a major event of EMT,22, 23 we examined the PI3K/Akt pathway in miR-19-expressing A549 and HCC827 cells. As shown in Supplementary Figure S3, miR-19 overexpression increased the phosphorylation of Akt in A549 and HCC827 cells, which was accompanied by a change of the phosphorylation of GSK-3β, a downstream target of Akt. These results suggest that miR-19 overexpression activated the Akt/GSK-3β pathway in A549 and HCC827 cells. In addition, increasing evidence supports that Stat3 has an important role in EMT of cancer cells.24, 25, 26 As shown in Supplementary Figure S3, overexpression of miR-19 also significantly increased the phosphorylation of Stat3 in A549 and HCC827 cells. These results suggest that miR-19 induced mesenchymal-like phenotypes and EMT-like cellular marker alterations in A549 and HCC827 cells through the PI3K/Akt pathway.
miR-19 overexpression induced epithelial–mesenchymal transition (EMT) of lung cancer cells. (a) The morphology of A549 and HCC827 cells expressing either the vector control or miR-19 was shown by phase contrast. (b) The relative messenger RNA levels of genes encoding E-cadherin, vimentin, fibronectin 1 (FN1), N-cadherin, and snail1 in A549 and HCC827 cells determined by quantitative reverse transcription-PCR. (c) The expression of the epithelial proteins E-cadherin, zona occludens 1 (ZO-1) and α-catenin, and the mesenchymal proteins vimentin, FN1, N-cadherin, and snail1 in the vector- and miR-19-expressing A549 and HCC827 cells detected by western blot. (d) Ectopic expression of miR-19 in A549 and HCC827 cells restored the disrupted F-actin stress fiber networks. Stress fibers (polymerized actin) and actin filaments were demonstrated by phalloidin staining (red). To observe F-actin cytoskeleton, fixed cells were incubated in 200 nM working stock of Acti-stain 670 phalloidin (Cytoskeleton). Cells were counterstained with DAPI (Sigma) and imaged using a confocal laser-scanning microscope (Olympus FV1000). The miR-19-expressing A549 and HCC827 cells showed clear stress fibers, whereas the vector-expressing A549 and HCC827 cells showed less stress fibers. DAPI, 4′,6-diamidino-2-phenylindole. *P<0.01.
miR-19 Overexpression Induced Global Expression Changes of Genes Related to EMT, Migration, and Metastasis in A549 Cells
The global gene expression changes induced by miR-19 overexpression were determined by comparing the gene expression profiles between miR-19- and vector-expressing A549 cells based on microarray (Figure 2). A total of 352 and 501 genes were significantly downregulated and upregulated, respectively, by miR-19 overexpression (Supplementary Figure S4; Supplementary Table S5).
miR-19 activated a subset of genes involved in EMT, migration, and metastasis of lung cancer cells. (a) Class comparison and hierarchical clustering of differentially expressed genes involved in EMT and migration between miR-19-expressing and vector-expressing A549 cells. In the cluster heatmap of genes involved in EMT, migration, and metastasis A1, A2, and A3 represented the total RNA (used in the microarray experiment) isolated from different generations of vector-expressing A549 cells and B1, B2, and B3 represented the total RNA (used in the microarray experiment) isolated from different generations of miR-19-expressing A549 cells. Genes with a fold change of higher than 2 and a P-value (t-test) of <0.05 were included in the analysis. Genes with increased and reduced expressions are shown in red and green, respectively. (b and c) Gene ontology (GO) (b) and KEGG pathway (c) analyses of up- and downregulated genes in miR-19- and vector-expressing A549 cells. Genes with expression changes of greater than twofold and P-values <0.05 were identified and classified using GO categories. (d) The messenger RNA levels of selected candidate genes from miR-19-expressing a549 cells were examined by quantitative reverse transcription-PCR. ECM, extracellular matrix; EMT, epithelial–mesenchymal transition; GO, Gene Ontology. *P<0.01.
The genes with significant express changes (Supplementary Figure S4) were submitted to the DAVID online tool (http://david.abcc.ncifcrf.gov/home.jsp) for GO annotation and pathway enrichment analysis. On the basis of GO terms, these genes were grouped into three major categories: biological process, cellular component, and molecular function. The GO terms representing biological processes related to EMT, migration, and metastasis are listed in Figure 2b and Supplementary Table S9. Among the 853 genes with significant expression changes, 206 genes were related to EMT, migration, and metastasis (upregulated: 134; downregulated: 72) (Figure 2a; Supplementary Tables S6 and S7). The GO terms representing cellular compartment and molecular function are listed in Supplementary Table S10.
Functional classification of the differentially expressed mRNA transcripts based on KEGG pathway analysis also demonstrated that the upregulated and downregulated genes are highly associated with focal adhesion, ECM–receptor interaction, regulation of actin cytoskeleton, cytokine–cytokine receptor interaction, TGF-beta signaling pathway, insulin signaling pathway, leukocyte transendothelial migration, MAPK signaling pathway, cell adhesion molecules, tight junction, and gap junction (Figure 2c; Supplementary Table S9).
To validate the microarray results, qRT-PCR was used to measure the expression of 18 genes (Figure 2d). The qRT-PCR results (Figure 2d) confirmed the expression changes of 13 genes (IGFBP3, ID2, PTEN, LPAR1, TGFB2, PIK3CD, TGFB1, PDGFA, TUBB2B, PDGFC, WNT5A, MMP10, and MMP1) identified by microarray (Supplementary Table S7). However, TWIST1 and TWIST2 without significant expression changes based on microarray were found to be significantly downregulated based on the qRT-PCR assay (Figure 2d). These inconsistent results may be explained by the low sensitivity of microarray compared with qRT-PCR in the detection of low-abundant transcripts.
On the basis of these results, miR-19 overexpression activated a number of EMT- and mobility-related genes in lung cancer cells, which is likely responsible for the switch on of the epithelial to mesenchymal gene expression program, as strongly supported by mesenchymal-like morphological conversion (Figure 1a), decreased cell adhesion (Figure 3a), and increased cell motility (Figure 3b) and invasion (Figure 3c).
miR-19 repressed the adhesion, and enhanced the motility and invasion of lung cancer cells undergoing epithelial–mesenchymal transition. (a) miR-19-expressing A549 cells exhibited reduced cell adhesion than the control cells. Vector- and miR-19-expressing A549 cells showed typical morphology of cell attachment (left). (b) The motility of miR-19-expressing A549 and HCC827 cells based on the transwell migration assay. (c) The invasion of miR-19-expressing A549 and HCC827 cells based on the Boyden chamber assay. *P<0.01 and #P<0.05.
Ectopic Expression of miR-19 Restored Disrupted Cytoskeleton in Lung Cancer Cells
The RhoA/ROCK-dependent pathway is involved in EMT and cytoskeletal signaling events that are crucial for cell motility. Cytoskeletal reorganization characterized by the formation of stress-fiber-bundling arrays is an important event in endocytosis, cell motility, and cancer cell invasion.19, 27, 28
As shown in Supplementary Figure S5 and Supplementary Tables S7 and S8, microarray analysis identified a number of cytoskeleton-related genes with significant upregulation (SPARC, TAGLN, CTGF, RHOU, RASGRP3, RHOB, PDGFA, ITGB6, ITGA5, PDGFC, RDX, and PALLD) and downregulation (ITGB4, ITGB8, MYO5C, MYO15B, VAV3, PLS1, ARHGAP26, and HOOK1) in the A549 cells undergoing EMT induced by miR-19. The GO terms related to cytoskeleton such as small GTPase-mediated signal transduction, regulation of cell shape, actin filament bundle formation, ruffle organization and biogenesis, barbed-end actin filament capping, and negative regulation of actin filament depolymerization are listed in Figure 2b and Supplementary Table S9. Functional classification of the differentially expressed mRNA transcripts based on KEGG pathway analysis also indicated that the some upregulated or downregulated genes were highly associated with the regulation of the actin cytoskeleton (Figure 2c; Supplementary Table S9). The GO terms representing cellular compartment (e.g., cytoskeleton, filopodium, lamellipodium, cortical actin cytoskeleton, ruffle membrane, actin cytoskeleton, ruffle, filamentous actin, cell projection, intermediate filament, and actin filament) and molecular function (e.g., actin binding, GTPase activity, and actin filament binding) related with cytoskeleton are listed in Supplementary Table S10.
The expression changes of the genes involved in cytoskeleton regulation prompted us to examine the status of stress fiber formation and polymerized actin in miR-19-expressing cells. Using phalloidin staining, we found that the disrupted F-actin stress fiber (polymerized actin) networks were restored in both miR-19-expressing A549 and miR-19-expressing HCC827 cells (Figure 1d). Therefore, ectopic expression of miR-19 promoted the formation of stress fibers in lung cancer cells.
These morphological alterations of the cytoskeleton are characteristics of EMT, in which epithelial cells lose polarity, cell–cell contacts, and cytoskeletal integrity, and acquire metastatic abilities.19, 27, 28 In summary, these results suggest that miR-19 overexpression induced EMT, and enhanced the mobility and invasion of lung cancer cells in vitro.
Decreased Cell Adhesion and Increased Cell Motility and Invasion of Lung Cancer Cells Undergoing EMT Induced by miR-19
Given the significant expression changes of genes involved in EMT and cell motility, we further investigated the adhesion, motility, and invasion abilities of miR-19-expressing lung cancer cells based on cell adhesion and migration assays. We found that miR-19-expressing A549 cells exhibited significantly reduced cell adhesion ability compared with control cells (Figure 3a), which was consistent with the expression changes of genes related to cell adhesion (e.g., E-cadherin and N-cadherin) (Figures 1b–d and 2b–d; Supplementary Tables S7). In addition, miR-19-expressing A549 and HCC827 cells exhibited mesenchymal-like phenotypes and significantly enhanced migration and invasion abilities compared with vector control cells (Figure 3b and c). Conversely, downregulation of endogeneous expression of miR-19 significantly reduced the migration and invasion abilities in A549 and HCC827 cells (Supplementary Figure S6). These results suggest that enforced expression of miR-19 in lung cancer cells triggered EMT, as well as reduced cell adhesion and increased cell migration and invasion.
PTEN is Involved in miR-19-Induced EMT, Migration and Invasion in Lung Cancer Cells
Generally, miRNAs regulate the expression of downstream gene(s) to exert their functions. Putative miR-19 targets involved in EMT and cell motility were predicted by searching a number of databases, such as microRNA.org, RNAhybrid, and miRWalk. Search in these databases revealed that PTEN was a potential target of miR-19. The 3′-UTR of PTEN mRNA contains a complementary site for the seed region of miR-19 (Figure 4a). To determine whether miR-19 regulates PTEN expression in lung cancer cells, we evaluated the expression of PTEN in lung cancer cell lines with miR-19 overexpression or miR-19 downregulation. The levels of PTEN in A549 and HCC827 cells were markedly reduced by ectopic expression of miR-19 (Figure 4c; Supplementary Figure S9B), while PTEN expression in A549 and HCC827 cells was significantly increased by silencing of miR-19 (Supplementary Figure S2). Furthermore, we evaluated the expression of miR-19a, miR-19b-1, and PTEN in available nine lung cancer cell lines (i.e., H1650, A549, HCC827, 95D, Spc-A-1, H358, H23, H446, and H460 cells) and the BEAS-2B cell line by qRT-PCR. qRT-PCR analysis revealed that the levels of miR-19a and miR-19b-1 were significantly upregulated when compared with BEAS-2B cells, and the expression level of PTEN was downregulated in lung cancer cell lines compared with BEAS-2B cells, suggesting a tendency that a high level of miR-19 expression is correlated with a low level of PTEN expression (Supplementary Figure S1). Therefore, there is a negative correlation between miR-19 and PTEN in lung cancer.
PTEN is involved in miR-19-induced EMT, cell migration, and invasion of lung cancer cells. (a and b) PTEN is a target gene of miR-19 in A549 cells based on the luciferase reporter assay. (c) Both miR-19 and siPTEN induced EMT-like cellular marker alterations, including reduced expression of the epithelial marker E-cadherin and increased expression of mesenchymal marker vimentin. (d) Both miR-19 and siPTEN enhanced the migration and invasion of lung cancer cells. Effects of miR-19 and siPTEN on cell migration and invasion were evaluated using the transwell chamber and Boyden chamber assays, respectively. Both ectopic expression of miR-19 and knockdown of PTEN promoted the migration and invasion of A549 and HCC827 cells. EMT, epithelial–mesenchymal transition; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; UTR, untranslated region. *P<0.01.
We further performed a luciferase reporter assay to determine whether miR-19 directly targets the 3′-UTR of PTEN in lung cancer cells. Transient transfection of wild-type PTEN-luc reporter with miR-19a mimics into A549 cells led to a significant decrease of luciferase activity compared with the mimics control (Figure 4b), whereas transient transfection of wild-type PTEN-luc reporter with miR-19a inhibitor led to a significant increase of luciferase activity compared with the inhibitor control (Figure 4b). These observations suggest that PTEN is a direct target of miR-19 in lung cancer cells.
To elucidate whether miR-19-induced EMT and miR-19-enhanced cell motility and invasion were mediated by PTEN, we silenced PTEN expression in A549 and HCC827 cells using siRNA. Our results demonstrated that similar to the ectopic miR-19 expression, siRNA-mediated knockdown of PTEN induced EMT, migration, and invasion of A549 and HCC827 cells (Figure 4c and d), suggesting that PTEN was involved in miR-19-induced EMT, cell migration, and invasion.
miR-19 Inhibited the Growth of Lung Cancer Cells Undergoing EMT Induced by miR-19 Expression
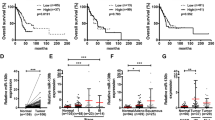
It has been reported that miR-17-92 gene cluster is frequently upregulated in human lung cancers, and improves the proliferation of cancer cells.6 However, we observed that A549 cells undergoing EMT induced by enforced expression of miR-19 exhibited low proliferative potential (Figure 5; Supplementary Figure S7), which was not consistent with previous studies.1, 5, 10, 29 Thus, this finding drove us to analyze the proliferation and cell-cycle progression of lung cancer cells undergoing EMT induced by miR-19. As demonstrated in Figure 5a, the results of the MTT assay demonstrated that miR-19 inhibited the growth of A549 cells undergoing EMT. In addition, as shown in a colony formation assay, miR-19-expressing A549 (Figure 5b) and HCC827 cells (Supplementary Figure S7) exhibited much less and smaller colonies than vector-expressing cells. Compared with LV-con, miR-19-expressing A549 cells exhibited increased percentage of cells in the G1 phase and decreased percentage of cells in the S-phase (Figure 5c). These results suggest that the growth-suppressive effect of miR-19 was partially due to a G1-phase arrest. In addition, we also observed that miR-19 suppressed the growth of tumors derived from A549 cells in nude mice (Figure 5d–f; Supplementary Figure S8).
Enforced expression of miR-19 inhibited the proliferation of lung cancer cells undergoing epithelial–mesenchymal transition. (a) Effects of miR-19 on the proliferation of A549 cells were analyzed by the MTT assay. (b) Representative pictures of colony formation assay of miR-19-expressing A549 cells. The colony formation ratio between miR-19-expressing A549 cells and vector-expressing A549 cells was determined to evaluate the effects of miR-19 on the proliferation of A549 cells. (c) Representative histograms of cell-cycle distribution of miR-19-expressing A549 cells. (d–f) miR-19 inhibited the growth of tumor caused by A549 cells in nude mice. (d) Representative picture of tumors in nude mice. (e) The growth curve of tumors in nude mice. (f) BrdU staining of transplanted tumors at 7 weeks after subcutaneous transplantation of miR-19-expressing A549 cells. Tumor weight and miR-19 expression in the xenografts are shown in Supplementary Figure S4. (g) Hierarchical clustering of differentially expressed genes related to cell cycle and cell proliferation in miR-19- and vector-expressing A549 cells. Right column lists the selected gene symbols, and the expression of genes related to cell cycle and cell proliferation are shown in the heatmap. (h) The expression of genes related to cell cycle and cell proliferation in miR-19-expressing A549 cells was analyzed by quantitative reverse transcription-PCR. BrdU, 5-bromo-2′-deoxyuridine; H&E, hematoxylin and eosin; MTT, 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-diphenytetrazoliumromide; OD, optical density. *P<0.01.
To identify genes involved in the growth inhibition, the mRNA levels of a number of cell-cycle regulators and proliferation-associated molecules in A549 cells were examined by microarray and qRT-PCR (Figure 5g and h; Supplementary Table S11). Microarray analysis revealed the upregulation of GNG4, MXD1, BCAT1, E2F7, and DAB2 and the downregulation of Betacellulin (BTC), Ets homologous factor, epiregulin (EREG), E2F transcription factor 8, and cell division cycle-associated 7-like (CDCA7L) (Figure 5g; Supplementary Table S11). In addition, qRT-PCR results demonstrated that the levels of E2F1, E2F2, CCNA2, and CCNB1 were significantly decreased, whereas the levels of tumor suppressor p21CIP1 was increased at least 3.5-fold by miR-19 overexpression (Figure 5h). Unexpectedly, miR-19 increased the mRNA level of CDK6 (Figure 5h; Supplementary Table S11), whereas the mRNA levels of cyclin-dependent kinase 2 and 4 (CDK2, CDK4) and cyclin D1 (CCND1) remained unaltered by miR-19 overexpression (Figure 5h). Therefore, overexpression of miR-19 in A549 cells undergoing EMT changed the expression of a number of cell-cycle regulators and cell growth-related proteins.
In summary, miR-19 inhibited the growth of lung cancer cells undergoing EMT through G1-phase arrest; however, the detailed mechanisms are still not clear.
miR-19 Promoted the Survival of Lung Cancer Cells Undergoing EMT Induced by miR-19 Expression
Cells undergoing EMT typically exhibit increased resistance to apoptosis.30, 31, 32, 33, 34 Therefore, we evaluated whether lung cancer cells undergoing EMT induced by miR-19 are more resistant to cell death. The microarray results revealed the upregulation of a number of anti-apoptosis genes such as BCL2-related protein A1 (BCL2A1), chemokine (C–C motif) ligand 2 (CCL2), tumor necrosis factor, alpha-induced protein 3 (TNFAIP3), matrix metallopeptidase 9, tumor protein p53-inducible nuclear protein 1 (TP53INP1), myeloid cell leukemia sequence 1 (BCL2-related, MCL1), baculoviral IAP repeat-containing 3 (BIRC3), and X-linked inhibitor of apoptosis (XIAP), and the downregulation of a number of pro-apoptosis genes such as TNFRSF11A, BTC, CD24, cell division cycle and apoptosis regulator 1 (CCAR1), and caspase 4 (Figure 6a; Supplementary Table S12). All GO terms representing biological processes listed in Figure 6b were related to apoptosis, including anti-apoptosis, negative regulation of apoptosis, induction of apoptosis, positive regulation of apoptosis, and regulation of apoptosis. Furthermore, the results of the qRT-PCR assay of selected genes including BCL2A1, XIAP, IL1RAP, IRAK2, TNFAIP3, IGFBP3, and TNFRSF11A were consistent with the microarray data (Figure 6c).
miR-19 improved the survival of cancer cells undergoing epithelial–mesenchymal transition. (a) Hierarchical clustering of differentially expressed genes related to apoptosis in miR-19- and vector-expressing A549 cells. Right column lists the selected gene symbols. The expression of genes related to anti-apoptosis and pro-apoptosis are shown in the heatmap. (b) GO analysis of up- and downregulated genes related to apoptosis in miR-19- and vector-expressing A549 cells. Genes with expression changes of higher than 1.7-fold and P-values <0.05 were identified and classified using GO categories. (c) The expression of genes related to anti-apoptosis and pro-apoptosis in miR-19-expressing A549 cells analyzed by quantitative reverse transcription-PCR. (d) miR-19 expression led to resistance to cell death of cancer cells induced by TNF-α. Vector- and miR-19-expressing cells were treated with TNF-α (5 ng/ml) after pretreatment with cycloheximide (0.5 μg/ml) for 30 min to prevent the induction of the survival protein NF-κB. (d-1) The cell cultures after 8 or 16 h of TNF-α treatment. (d-2) The activities of the death receptor-specific caspase-8 and effector caspase-3. The low activities of both caspases in miR-19-expressing cells indicated the healthy appearance observed in d-1. GO, Gene Ontology; TNF-α, tumor necrosis factor alpha. *P<0.01.
Next, we checked whether miR-19-expressing cells were resistant to apoptosis induced by proapoptotic signal such as TNF-α. Most vector-expressing A549 cells died within 16 h after TNF-α treatment; however, most miR-19-expressing A549 cells survived with TNF-α treatment (Figures 6d-1). Caspase-8 is specifically recruited to death receptors and autoactivated to initiate the apoptotic pathway. Accordingly, we found that TNF-α treatment activated caspase-8 in vector-expressing A549 cells (Figure 6d-2). An approximately threefold decrease in the activity of caspase-8 was observed in miR-19-expressing A549 cells after 16 h of TNF-α treatment compared with that of vector-expressing A549 cells (Figure 6d-2). These results suggest that the death of vector-expressing A549 cells was induced by activation of the apoptotic pathway (Figure 6d-2). As expected, the activity of the initiator caspase-8 correlated with that of caspase-3 (the effector) (Figure 6d-2), suggesting that miR-19 overexpression prevented TNF-α-induced death of A549 cells undergoing EMT. Therefore, miR-19 enhanced the antiapoptotic ability of lung cancer cells undergoing EMT.
DISCUSSION
Invasion and metastasis, the most significant features of cancers, are commonly used as prognostic factors to evaluate the survival of cancer patients. Therefore, understanding the molecular mechanisms of cancer invasion and metastasis may lead to the identification of novel therapy approaches against cancers.19, 27, 28 Given that EMT has an important role in cancer invasion and metastasis, we investigated the molecular mechanisms of EMT in lung cancer cells, which may allow us to identify key factors involved in cancer invasion and metastasis.
It has been reported that miR-19 was dysregulated in various cancers, and miR-19 upregulation strongly correlated with gain of invasive and metastatic properties and poor prognosis of patients with NSCLC,11, 12, 13 gastric cancer,14 and esophageal squamous cell carcinoma.15 Moreover, the expression of miR-19 was significantly higher in the less-invasive breast cancer MCF7 cells than the more-invasive MDAMB-231 cells, and the expression of a positive regulator of tumor angiogenesis and metastasis was negatively regulated by miR-19 in breast cancer cells,35 suggesting that miR-19 inhibited the metastasis of breast cancer. In osteosarcoma cells, the expression of miR-19a was higher in metastatic LM7 cells with low expression of Fas than the parental nonmetastatic SAOS-2 cells with high expression of Fas.36 Recent studies have shown that miR-19a and miR-19b-1 promoted the migration of cervical carcinoma cells16 and colon cancer cells.17 In addition, it has been reported that miR-19a/b facilitated the migration, invasion, and metastasis of gastric cancer cells by targeting the tumour suppressor MXD1.14 Furthermore, Fan et al.18 reported that miR-19b promoted the migration, invasion, and metastasis of Hela, MCF7, and Huh7 cells by targeting TP53. These studies strongly support that miR-19 has a central role in cancer invasion and metastasis by regulating different targets; however, the underlying mechanisms are largely unknown.
EMT is a characteristic event in the invasion and metastasis of various cancers.19, 20 Increased evidence shows that miRNAs are closely associated with EMT, invasion, and metastasis of cancer.37, 38, 39, 40, 41 For e.g., let-7, miR-30a, miR-125a-5p, miR-126, miR-183, miR-200, miR-21, miR-221, miR-222, and miR-328 have important roles in EMT, invasion, and metastasis of lung cancer.42, 43, 44, 45, 46, 47 In this study, we found that enforced expression of miR-19 (i.e., miR-19a and miR-19b-1) in lung cancer cells A549 and HCC827 cells triggered EMT, as shown by mesenchymal-like morphological changes, downregulation of epithelial proteins (e.g., E-cadherin, ZO-1, and α-catenin), upregulation of mesenchymal proteins (e.g., vimentin, FN1, N-cadherin, or snail1), formation of stress fibers, reduced cell adhesion, and enhanced cell migration and invasion. In addition, DNA microarray results also highlighted the expression of genes involved in EMT, migration, and metastasis in miR-19-expressing A549 cells. These observations suggest that miR-19 induced EMT, which contributed to the invasion and metastasis of lung cancers.
Actin polymerization and depolymerization, which allow cells to protrude at the anterior front, are prominent events in cell movements to designated directions.48 Concurrently, consecutive actomyosin contraction and separation from the posterior end further drive cells to move.48 Actin polymerization is also involved in the formation of stress fibers.48 Therefore, dysfunction of actin polymerization leads to reduced motility and invasion of cells.48 In the present study, our results demonstrated that exogenous expression of miR-19 in A549 and HCC827 cancer cells restored the formation of stress fibers, suggesting that miR-19 regulated lung cancer cell migration and invasion through promoting the formation of stress fibers.
We also provided evidence supporting that PTEN was a potentially functional target of miR-19 and was involved in miR-19-induced EMT. Our results showed that miR-19 directly bound to the complementary sites of 3′-UTR of PTEN (Figure 4a), and markedly decreased the protein level of PTEN in lung cancer cells (Figure 4c; Supplementary Figure S9B). It has been reported that loss of function or downregulation of PTEN resulted in tumor metastasis, and more aggressive growth behaviors of tumor, and/or poor prognostic phenotypes.42, 49, 50, 51, 52, 53, 54, 55, 56, 57 In addition, miR-19 upregulation was observed in various cancers including lung cancer, and correlated with tumor metastasis and/or poor prognosis of cancer patients.11, 12, 13, 14, 15, 18 Given that miRNAs primarily function through inhibiting the translation of their mRNA targets, we propose that miR-19 might be a negative regulator for PTEN, which has been reported by other researchers.10, 29, 58, 59 In addition, miR-19 increased the resistance to apoptosis of mouse B-cell lymphoma,10, 29 and chemoresistance of human breast cancer58 and gastric cancer59 through directly repressing PTEN expression. These studies further support our hypothesis that miR-19 inhibits the expression of PTEN. Next, we continued to determine whether miR-19 positively modulates EMT, migration, and invasiveness of cancer cells in vitro via regulation of PTEN. Increasing evidence supports that inactivation or downregulation of the tumor suppressor PTEN can trigger EMT of cancer cells,60, 61, 62, 63 which then promoted the invasion and metastasis of various cancers including lung cancer.42, 49, 50, 51, 52, 53, 54, 55, 56, 57 It has been reported that PTEN negatively regulated the PI3K/Akt pathway through the dephosphorylation of PI(3,4,5)P3, and ultimately participated in the regulation of cell cycle, proliferation, apoptosis, adhesion, and EMT during cancer progress.63 In the present study, we found that PTEN silence through RNA interference mimicked miR-19-induced EMT, and promoted the migration and invasion of tumor cells, which were similar as the results caused by miR-19 overexpression. Taken together, miR-19 triggered EMT and improved the invasion and metastasis of cancer cells through inhibiting the expression of PTEN.
Interestingly, we found that miR-19-overexpressing lung cancer cells exhibited reduced proliferation and enhanced survival during the process of EMT. It seems that the proliferation inhibition of miR-19 in lung cancer cells is inconsistent with its oncogene role.1, 5, 6, 10, 16, 17, 29 It has been reported that EMT contributed to the invasion and migration of tumor cells, and EMT was attributed to the metastatic dissemination and the resistance of tumor cells to radiotherapy, chemotherapy, and apoptosis.19, 32, 39 Tumor cells undergoing EMT are typically found at the edge of tumors, account for a small proportion of primary cancer, and have low proliferative rates. Strong evidence suggests that reduced proliferation cooperates with EMT to drive cancer cells to reach their new destinations before efficient proliferation.30, 31, 32, 34, 64, 65 In addition, the products of EMT-induced genes, such as Snail1,34 ZEB2/SIP1,65 and Y-box-binding protein-1 (YB-1)32, 64 exhibited strong anti-proliferative activities by inhibiting the cell cycle of tumor cells undergoing EMT, which is consistent with our results. Previous studies and our results suggest that upregulation of cell proliferation is important for the initiation and maintenance of primary tumors, while growth inhibition is crucial for the invasion and migration of tumor cells.31, 32, 34, 64, 65 Taken together, the oncogene miR-19 can promote the proliferation of tumor cells during the initiation and maintenance of primary tumors, and induce EMT of tumor cells to enhance tumor invasion and metastasis, suggesting functional transition of miR-19 at different stages of tumor development.
Resistance to apoptosis confers a selective advantage to invasion and migration of cells during both embryonic development and tumor metastasis.30, 31, 32, 33, 34 In addition, tumor cells undergoing EMT are more resistant to apoptosis.30, 31, 32, 33, 34 Reduced proliferation may increase the resistance to apoptosis of cancer cells undergoing invasion and metastasis.32 Antiapoptotic activity is another common feature of the genes induced by EMT, such as Snail1,30, 34, 66 ZEB2/SIP1,31, 39, 67 YB-132, 64 and miR-19 (the present study). Therefore, resistance to apoptosis conferred by miR-19 provides a selective advantage to the invasion and migration of cancer cells. As shown in Supplementary Figure S10, overexpression of miR-19 led to the loss of epithelial markers, the expression of mesenchymal markers, changes of cell shape, and the acquisition of motility and invasive properties, and apoptosis resistance of lung cancer cells.
In summary, we demonstrated that upregulation of miR-19 induced EMT and promoted the invasion of lung cancer cells, but the roles of miR-19 in the metastasis of lung cancer cells and the underlying mechanisms remain to be well investigated in the future. In addition, enforced expression of miR-19 resulted in reduced proliferation of tumor cells undergoing EMT, which has not yet been reported in other human cancers. These slow-growing, apoptosis-resistant, and highly invasive lung cancer cells are likely insensitive to therapeutic interventions that primarily target highly proliferating cells. The underlying mechanisms remain to be fully explored in the future. Interestingly, miR-19-expressing lung cancer cells appear to possess several stem cell features, including low proliferation rates and upregulation of the stem cell markers CD133 and Bmi-1 (data not shown). Thus, further investigations are necessary to determine whether miR-19 reexpression and EMT inducers are associated with cancer stem cell phenotypes.39
References
Croce CM . Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–714.
Garzon R, Calin GA, Croce CM . MicroRNAs in cancer. Annu Rev Med 2009;60:167–179.
Mendell JT . miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008;133:217–222.
Olive V, Jiang I, He L . mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010;42:1348–1354.
van Haaften G, Agami R . Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev 2010;24:1–4.
Hayashita Y, Osada H, Tatematsu Y et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628–9632.
Liu M, Wang Z, Yang S et al. TNF-alpha is a novel target of miR-19a. Int J Oncol 2011;38:1013–1022.
Connolly E, Melegari M, Landgraf P et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol 2008;173:856–864.
Sang L, Roberts JM, Coller HA . Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med 2010;16:17–26.
Olive V, Bennett MJ, Walker JC et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev 2009;23:2839–2849.
Navarro A, Marrades RM, Vinolas N et al. MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology 2009;76:162–169.
Wu C, Cao Y, He Z et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med 2014;232:85–95.
Lin Q, Chen T, Lin Q et al. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol 2013;107:767–771.
Wu Q, Yang Z, An Y et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis 2014;5:e1144.
Xu XL, Jiang YH, Feng JG et al. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg 2014;97:1037–1045.
Zhang J, Xiao Z, Lai D et al. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. Br J Cancer 2012;107:352–359.
Xu XM, Wang XB, Chen MM et al. MicroRNA-19a and -19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett 2012;322:148–158.
Fan Y, Yin S, Hao Y et al. miR-19b promotes tumor growth and metastasis via targeting TP53. RNA 2014;20:765–772.
Thiery JP, Sleeman JP . Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131–142.
Christofori G . New signals from the invasive front. Nature 2006;441:444–450.
Shan SW, Lee DY, Deng Z et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 2009;11:1031–1038.
Larue L, Bellacosa A . Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene 2005;24:7443–7454.
Bakin AV, Tomlinson AK, Bhowmick NA et al. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem 2000;275:36803–36810.
Liu RY, Zeng Y, Lei Z et al. JAK/STAT3 signaling is required for TGF-beta-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol 2014;44:1643–1651.
Cho KH, Jeong KJ, Shin SC et al. STAT3 mediates TGF-beta1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett 2013;336:167–173.
Hu QD, Chen W, Yan TL et al. NSC 74859 enhances doxorubicin cytotoxicity via inhibition of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Lett 2012;325:207–213.
Kalluri R, Weinberg RA . The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420–1428.
Yilmaz M, Christofori G . EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009;28:15–33.
Mu P, Han YC, Betel D et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 2009;23:2806–2811.
Barrallo-Gimeno A, Nieto MA . The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 2005;132:3151–3161.
Browne G, Sayan AE, Tulchinsky E . ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle 2010;9:886–891.
Evdokimova V, Tognon C, Ng T et al. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 2009;8:2901–2906.
Schickel R, Park SM, Murmann AE et al. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell 2010;38:908–915.
Vega S, Morales AV, Ocana OH et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 2004;18:1131–1143.
Zhang X, Yu H, Lou JR et al. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J Biol Chem 2011;286:1429–1435.
Huang G, Nishimoto K, Zhou Z et al. miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res 2012;72:908–916.
Zhao X, Lu Y, Nie Y et al. MicroRNAs as critical regulators involved in regulating epithelial- mesenchymal transition. Curr Cancer Drug Targets 2013;13:935–944.
Sreekumar R, Sayan BS, Mirnezami AH et al. MicroRNA control of invasion and metastasis pathways. Front Genet 2011;2:58.
Polyak K, Weinberg RA . Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265–273.
Nicoloso MS, Spizzo R, Shimizu M et al. MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 2009;9:293–302.
Bullock MD, Sayan AE, Packham GK et al. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell 2012;104:3–12.
Zhang JG, Wang JJ, Zhao F et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 2010;411:846–852.
Yang Y, Ahn YH, Gibbons DL et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest 2011;121:1373–1385.
Kumarswamy R, Mudduluru G, Ceppi P et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer 2012;130:2044–2053.
Du L, Pertsemlidis A . microRNAs and lung cancer: tumors and 22-mers. Cancer Metastasis Rev 2010;29:109–122.
Cherni I, Weiss GJ . miRNAs in lung cancer: large roles for small players. Future Oncol 2011;7:1045–1055.
Arora S, Ranade AR, Tran NL et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer 2011;129:2621–2631.
Zheng F, Liao YJ, Cai MY et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012;61:278–289.
Abate-Shen C, Banach-Petrosky WA, Sun X et al. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res 2003;63:3886–3890.
Akca H, Demiray A, Tokgun O et al. Invasiveness and anchorage independent growth ability augmented by PTEN inactivation through the PI3K/AKT/NFkB pathway in lung cancer cells. Lung cancer 2011;73:302–309.
Chung MJ, Jung SH, Lee BJ et al. Inactivation of the PTEN gene protein product is associated with the invasiveness and metastasis, but not angiogenesis, of breast cancer. Pathol Int 2004;54:10–15.
Damsky WE, Curley DP, Santhanakrishnan M et al. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 2011;20:741–754.
Dankort D, Curley DP, Cartlidge RA et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009;41:544–552.
Ratnacaram CK, Teletin M, Jiang M et al. Temporally controlled ablation of PTEN in adult mouse prostate epithelium generates a model of invasive prostatic adenocarcinoma. Proc Natl Acad Sci USA 2008;105:2521–2526.
Sawai H, Yasuda A, Ochi N et al. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol 2008;8:56.
Shih MC, Chen JY, Wu YC et al. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene 2012;31:2389–2400.
Sze KM, Wong KL, Chu GK et al. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology 2011;53:1558–1569.
Liang Z, Li Y, Huang K et al. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res 2011;28:3091–3100.
Wang F, Li T, Zhang B et al. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun 2013;434:688–694.
Bowen KA, Doan HQ, Zhou BP et al. PTEN loss induces epithelial—mesenchymal transition in human colon cancer cells. Anticancer Res 2009;29:4439–4449.
Mulholland DJ, Kobayashi N, Ruscetti M et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 2012;72:1878–1889.
Wang H, Quah SY, Dong JM et al. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res 2007;67:2922–2926.
Song LB, Li J, Liao WT et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009;119:3626–3636.
Evdokimova V, Tognon C, Ng T et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 2009;15:402–415.
Mejlvang J, Kriajevska M, Vandewalle C et al. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol Biol Cell 2007;18:4615–4624.
Franco DL, Mainez J, Vega S et al. Snail1 suppresses TGF-beta-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J Cell Sci 2010;123 (Pt 20):3467–3477.
Sayan AE, Griffiths TR, Pal R et al. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci USA 2009;106:14884–14889.
Acknowledgements
We thank Prof. Andrea Ventura (Memorial Sloan Kettering Cancer Center) and Dr Jeng-Shin Lee (Harvard Gene Therapy Initiative, Harvard Medical School) for generously providing plasmids. This work was supported by the National Natural Science Foundation of China (Grant Nos 81172587 and 81372896 to DX; Grant No. 81101535 to D-JL; Grant No. 61427807 to W-HH), the Natural Science Foundation of Guangdong Province of China (Grant No. 2014A030313294 to DX; Grant No. 9151063101000015 to J-SJ), the Science and Technology Planning Project of Guangdong Province of China (Grant Nos 2009B060300008 and 2013B060300013 to DX) and the major project funding was from the R & D institutions of Fujain (2012I2004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Overexpression of miR-19 is associated with poor prognosis in lung cancer. This study reveals that miR-19 triggers epithelial–mesenchymal transition (EMT) in lung cancer cells, which plays an important role in invasion and migration. Furthermore, lung cancer cells undergoing EMT induced by miR-19 demonstrate enhanced resistance to apoptosis growth inhibition.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, J., Yang, S., Yan, W. et al. MicroRNA-19 triggers epithelial–mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest 95, 1056–1070 (2015). https://doi.org/10.1038/labinvest.2015.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.76
This article is cited by
-
Oncogenic role of microRNA-19b-3p-mediated SOCS3 in glioma through activation of JAK-STAT pathway
Metabolic Brain Disease (2023)
-
Rs7853346 Polymorphism in lncRNA-PTENP1 and rs1799864 Polymorphism in CCR2 are Associated with Radiotherapy-Induced Cognitive Impairment in Subjects with Glioma Via Regulating PTENP1/miR-19b/CCR2 Signaling Pathway
Biochemical Genetics (2022)
-
miR-26a promotes hepatocellular carcinoma invasion and metastasis by inhibiting PTEN and inhibits cell growth by repressing EZH2
Laboratory Investigation (2019)
-
miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer
Molecular Cancer (2018)
-
Development of Novel Monoclonal Antibodies for Evaluation of Transmembrane Prostate Androgen-Induced Protein 1 (TMEPAI) Expression Patterns in Gastric Cancer
Pathology & Oncology Research (2018)