Abstract
Archived tumor specimens, particularly those collected by large cooperative groups and trials, provide a wealth of material for post hoc clinical investigation. As these tissues are rigorously collected and preserved for many decades, subsequent use of the specimens to answer clinical questions must rely on the assumption that expression and detection of target biomarkers are not degraded with time. To test this assumption, we measured the expression of estrogen receptor (ER), human epidermal growth receptor 2 (HER2), and Ki67 in human breast carcinoma using quantitative immunofluorescence (QIF) in a series of formalin-fixed paraffin-embedded (FFPE) tissues from 1295 individual patients preserved for 7 to 53 years in four cohorts on tissue microarrays. Protein expression was measured using the automated quantitative analysis method for QIF. Change in quantitative protein expression over time was estimated in positive cases using both Pearson’s correlation and a polynomial regression analysis with a random effects model. The average signal decreased with preservation time for all biomarkers measured. For ER and HER2, there was an average of 10% signal loss after 9.9 years and 8.5 years, respectively, compared with the most recent tissue. Detection of Ki67 expression was lost more rapidly, with 10% signal loss in just 4.5 years. Overall, these results demonstrate the need for adjustment of tissue age when studying FFPE biospecimens. The rate of antigenicity loss is biomarker specific and should be considered as an important variable for studies using archived tissues.
Similar content being viewed by others
Main
Investigation of prognostic markers and predictive biomarkers is commonly performed using archived tissue specimens. Cooperative groups maintain archived formalin-fixed, paraffin-embedded (FFPE) tissue from their trials, and hospitals maintain archives of excised tumor specimens that can be associated with institutional records providing long-term follow-up. These archives that combine large volumes of samples with outcome data offer a valuable resource for validation of existing biomarkers and discovery of new biomarkers. They facilitate retrospective studies and even higher level of evidence prospective-retrospective studies. These ‘prospective-retrospective’ studies benefit from the rigorous inclusion criteria, randomization, intervention, and endpoint collection of a prospective trial, while being utilized in a retrospective manner for tissue analysis as new questions arise.1 For example, the North Central Cancer Treatment Group Trial N9831, a randomized, phase III trial demonstrating clinical benefit with trastuzumab in early stage HER2-positive breast cancers,2 has been used in this context. Perez et al3 used a tissue microarray (TMA) of ~800 patients to investigate the potential association between C-MYC alterations and resistance to trastuzumab therapy.
Similarly, well-preserved tissue banks will be important to validate clinical evidence from these studies in the future, as new techniques and standards come into practice. For example, concerns about local laboratory HER2 testing in three earlier clinical trials using trastuzumab in metastatic breast cancer (H0648g, H0649g, and H0650g) prompted a retrospective validation of all patients’ HER2 expression using archived specimens.4 Based on these and many other successes, recent groups have recommended that all prospective clinical trials include formalized biospecimen banking for use in this context.5 Cooperative groups that participate in clinical trials can use the prospective-retrospective model to share both aspects of investigation—subject enrollment, treatment, and analysis—as well as post hoc investigations of new questions.
One challenge of archived specimens is the limited amount of preserved tissue available for multiple investigations. After required materials are collected and used for clinical and diagnostic procedures, uses of the remaining tissue must be carefully chosen. TMAs offer a significant advantage to allow multiple uses, amplifying a single specimen as much as 1000-fold.6 Furthermore, TMAs allow side-by-side analysis of multiple cohorts with the same experimental and treatment conditions.
Use of archived tissues, including both TMAs and whole sections, to validate biomarkers with clinical utility relies on the assumption that expression and measurement of the target of interest is comparable to tissue collected in the present day. Many patient cohorts, both prospectively and retrospectively collected, span several years of treatment, and with follow-up of 10, 20 years or more many of these cohorts are decades old. Thus, the validity of these results should be assessed before those results are applied to current clinical practice. In particular, the preservation of biomarker expression and integrity is unknown over long periods of time. After sectioning, significant loss of immunoreactivity in tissue slides has been documented in weeks to months, despite optimal storage conditions including paraffin coating and nitrogen desiccation.7, 8 In contrast to cut and sectioned tissue, protein expression is considered well-maintained in paraffin-embedded tissue blocks for many years.9 Theoretically, unsectioned tissue in paraffin blocks should be less exposed to degrading effects, though the persistence and reliability of antigenicity in these tissues over many years remain poorly characterized.
The assessment of this problem can be a challenge since the effect may be subtle and possibly undetectable with conventional subjective or non-standardized IHC measurement or with cohorts that span only a few years. Here we use tissue specimens spanning over 40 years and quantitative assessment of protein expression to attempt to detect changes in antigenicity of archival tissue stored as paraffin blocks. Specifically, we analyzed protein expression of three clinically-relevant biomarkers, estrogen receptor (ER), human epidermal growth receptor 2 (HER2), and Ki67, as well as the epithelial marker cytokeratin (CK), in archived breast carcinoma tissue between 7- and 53-year old.
MATERIALS AND METHODS
Tissue Selection and Preparation
Invasive breast carcinoma tissue was obtained from the archives of the Pathology Department of Yale University (New Haven, CT). Cohorts represent serial collection of breast cancer specimens received by our surgical pathology suite. All specimens were collected from patients at Yale New Haven Hospital between 1961 and 2006, and each tumor was FFPE at the time of surgery according to standard operating procedures. Tissues were prepared for high-throughput assays by creating TMAs, which include 0.6-mm cores from patient tumors, as well as tissue and cell line controls as previously described.10 Four cohorts of patients were selected to use the widest breadth of tissue ages, and included 1330 individual tumor samples. Yale TMA-49, TMA-201, TMA-37, and TMA-128 included patient tissue samples collected from 1961 to 1983, 1975 to 2003, 1991 to 1997, and 2002 to 2006, respectively (Figure 1). For comparative assays for each target, slides from each cohort were freshly cut from the paraffin-embedded TMA block and stained on the same day to eliminate the variable of diminished expression as a function of time after sectioning.
Fluorescence Immunohistochemistry
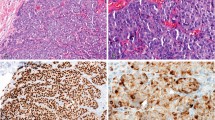
Protein expression was detected by fluorescent immunohistochemistry. All four TMAs were stained side-by-side for each target. Slides were deparaffinized by baking at 60° for 30 min followed by two xylene treatments for 20 min each. After rehydration with decreasing ethanol concentrations, antigen retrieval was performed using a Lab Vision PT Module with citrate buffer (pH 6.0) at 97° for 20 min. Permeabilization with 0.3% H2O2 in methanol was then followed by non-specific antibody blocking with 0.3% bovine serum albumin (BSA) in 0.1 M Tris-buffered saline (TBS, pH 8) for 30 min at room temperature.
The primary antibodies included anti-ER (Thermo Scientific SP1, rabbit monoclonal at 1:1000) or anti-HER2 (Dako A0458, rabbit polyclonal at 1:10 000), or anti-Ki67 (Dako MIB-1 mouse monoclonal at 1:100) diluted in 0.3% BSA/TBS. For ER and HER2 assays, each slide was also treated with mouse anti-human CK (Dako, 1:100) to identify epithelial tumor cells (rabbit anti-cow CK was used with anti-Ki67). Slides were incubated with primary antibodies overnight at 4 °C (ER and HER2) or for 1 h at room temperature (Ki67). After washing, secondary antibody conjugated to a Cy3 fluorophore (AlexaFlour 546 goat anti-rabbit or anti-mouse, 1:100, Life Technologies) diluted in an anti-mouse or -rabbit secondary conjugated to horseradish peroxidase (EnVision, Dako). To visualize and amplify the target signal, slides were then washed and incubated with Cy5-conjugated tyramide (1:500, PerkinElmer) for 10 min. Prolong Gold mounting medium containing DAPI (4,6-diamidino-2-phenylindole; Life Technologies) was used to visualize tissue nuclei.
Automated Quantitative Immunofluorescence
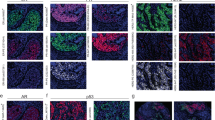
Each tissue core sample on the TMA was captured by a HistoRX PM2000 microscope in three channels: nuclear DAPI, CK-C3, and target-Cy5. Automated quantitative analysis (AQUA) allows measurement of protein concentration within subcellular compartments.10 The CK pattern was used to distinguish tumor cells from stromal components and create an epithelial compartment. A nuclear compartment is defined by the DAPI signal within the epithelial compartment, to include only tumor cell nuclei. The final AQUA score represents the target compartment pixel intensities divided by the tumor compartment (CK) or nuclear compartment (DAPI) pixels.
As ER and Ki67 are nuclear proteins, the nuclear compartment was used for target scoring, while HER2, a membranous protein, was scored in the epithelial compartment. Finally, we measured the expression of CK in the epithelial compartment run with the anti-HER2 antibody to have a positive control target across all years.
Data Analysis
Tissue specimens randomly lost from the TMA or with missing year of tissue fixation were excluded from analysis. For 23 tumors with duplicate cores, the max score was used. Since our goal was to detect loss of antigenicity, negative cases were also excluded from analysis. To determine the cut-point to define negative cases, without bias from year of collection, we used expected population cut points from the literature. According to population data from a large, unrestricted breast cancer cohort,11 we selected the highest 69% of ER cases, the highest 13% of HER2 cases, and the highest 33% of Ki67 cases in each of the three cohorts to ensure the comparison of biomarker-positive tissue. All cases stained for CK were included. In the exploratory analysis, we used scatter plots to demonstrate the relationship between AQUA scores and the year of tissue fixation for each biomarker. Pearson’s correlation coefficient with the P-value from the two-sided test against zero was used to assess correlation of AQUA scores with tissue year. Polynomial regression analysis with backward variable selection was used to model the relationship between tissue age and log of the AQUA scores. The log transformation was applied based on the inspection of the normality assumption using Anderson–Darling’s test and residual plot, where the AQUA scores were highly skewed to the right in the raw scale but not in the log scale. A random effect for cohort was included in the regression analysis to adjust the potential unknown systematic error (bias) associated with the cohort. An estimated regression coefficient with the type III P-value<0.05 was regarded as statistically significant. Point prediction and the corresponding 95% point-wise prediction interval were computed and plotted in the range of the observed tissue age. All statistical analyses were conducted using Statistical Analysis System (SAS) software, version 9.4 (SAS Institute, Cary, NC).
RESULTS
Available Tissue and AQUA Score Distributions
The four cohorts selected for AQUA analysis included tumor specimens from 1295 individuals (or 829 with tissue) obtained and fixed between 1961 and 2006. Median tissue age was 28 years (IQR 14–40 years), correlating to tissue fixed in 1986. Tissue and signal were sufficient for scoring for 630 patients in the ER assay, 603 patients in the HER2 assay, 707 patients in the Ki67 assay, and 647 in the CK assay. The distribution of these cohorts by age and antigen is schematically demonstrated in Figure 1. ER scores had a distribution, from 35 to 17 098 in the nuclear compartment. Among the top 69% of scores in each cohort (n=435), the minimum score was 324. The range of HER2 scores was from 74 to 14 729 in the epithelial compartment defined by CK staining, with 78 cases included in the top 13% from each cohort, minimum 1037. Ki67 expression ranged from 62 to 15 767 in the nuclear compartment, and minimum score of the top 33% of cases was 273 (n=233).
Correlation Between Expression Level and Tissue Age in Positive Cases
Pearson’s correlation between the year of diagnosis and AQUA score was completed for all proteins. Distributions by year of tissue fixation are shown in scatter plots (Figure 2a–d). For ER cases, there was a positive correlation with year and AQUA score (r2=0.06, P<0.0001). According to this exploratory analysis, the predicted ER AQUA score decreases by 94 units on average each year since the tissue sample collection is consistent with a 10% decrease in average signal after 9.9 years. HER2 highest cases also showed lower scores in older tissue specimens (r2=0.11, P=0.0034). The slope indicates a decrease of 69 AQUA units every year with a 10% loss of signal in about 8.5 years. Ki67 cases above the cutoff had the strongest correlation with r2=0.22 (P<0.0001) and a predicted decrease of 66 AQUA units per year. Ki67 demonstrated the most rapid proportional loss of signal over time, with a 10% decrease in AQUA signal after only 4.5 years. For comparison, positive CK scores were analyzed for all cases, and demonstrated a weak, but significant, correlation with tissue age (r2=0.03, P<0.0001) and a slope of 131 AQUA units per year. In contrast to the other biomarkers, CK maintained 90% of its AQUA signal up to 22.5 years after specimen collection and fixation. Note that this study was done in 2014 and the newest tissues were from 2006, so we make the assumption that the signal loss seen in the initial period (between 2014 and 2006) is linear with respect to the data collected during the study years.
The distribution of scores for each biomarker as a function of tissue age after omitting the fraction of expected negative cases. (a) ER, (b) HER2, (c) Ki67 and (d) cytokeratin. The fraction of positive cases is shown by percentage beneath the biomarker in the title. The regression value and P-value are presented in the insets. Au, arbitrary unit.
Modeling AQUA Scores with the Tissue Age
Consistent with the correlations above, according to the random effects model the logarithmic AQUA score for all biomarkers decreased with tissue age. The predicted AQUA score for both ER and HER2 expression levels decreased with age (P<0.001 and P=0.004, respectively, Figure 3a and b). The linear relationship of the predicted AQUA scores for ER and HER2 indicates a steady decline in tissue expression over time. In contrast, for Ki67, a quadratic equation was a better model for the change in AQUA score over time (p(x2)=0.0387, Figure 3c). This relationship suggests that the greatest loss of signal for Ki67 occurs in the first decades after tissue fixation, and the loss of expression plateaus after about 30 years. The equation for predicted expression of CK based on tissue age was also linear (Figure 3d), consistent with a consistent loss of signal over time.
DISCUSSION
Using quantitative measurement of protein expression, we observed a decrease in signal detection in older tissues across five decades in four proteins: ER, HER2, Ki67, and CK. These results represent unsectioned tumor specimens stored in paraffin-embedded blocks and do not represent tumor specimens stored as sectioned tissue on glass slides (these degrade much more rapidly). Consistent with prior studies,9 positive cases of ER and HER2, and maintenance of CK expression, can be detected in tissue samples from over 50 years ago. However, the magnitude of the signal appears to diminish over time. Thus, it is likely that some cases are no longer detectable as positive although they might have been above the threshold had they been measured years ago. For Ki67, the loss of immunoreactivity is most marked with no high-expressing cases detected prior to the 1980s, indicating that some biomarkers are more susceptible to antigen degradation than others. Wester et al12 previously demonstrated different rates of antigen loss in paraffin sections, including particularly significant decrease with Ki67 antibody MIB-1. In addition, the exponential relationship between tissue age and Ki67 signal indicates a prominent decrease in detectable signal in the first 30 years that subsequently levels off, in contrast to a slower, steady decline for other biomarkers.
Given the loss of antigenicity and our calculations of loss per year, tentative recommendations can be made regarding when antigen degradation may affect results. As our coefficient of variation for quantitative assessment of expression of protein by AQUA is ~10%, when the age has caused the AQUA score to decrease by >10% we need to adjust for this or avoid the use of tissue sample that is older than that age. ER protein expression detected by AQUA was diminished by an average of 10% after about 10 years, and that of HER2 after 8.5 years. Ki67 signal was lost much more quickly, with a 10% signal loss in only 4.5 years after fixation. Fortunately, CK expression persisted for over 20 years with no more than 10% signal loss. These results demonstrate that loss of antigenicity is target specific, and certain proteins may be less well-preserved than others.
Several investigations of pre-analytic variables have demonstrated loss of protein expression in tissue sections due to effects of air, humidity, temperature, and fixatives, leading to mechanisms of protein damage or modification including oxidation, cross-linking, and denaturation.8, 13, 14 Whether these processes affect unsectioned tissue, and to what degree they cause signal loss is not well-described. The data here suggest that signal degradation does occur in this setting. To minimize other explanations for antigen loss, we used TMAs and slides cut and stained on the same day to decrease specimen and inter-array variability and to minimize any effects of slide storage, as previously described.8
While we believe that the effects we have described are a function of tissue aging within the paraffin block, there are limitations to this study. Perhaps most significant is the lack of fresh or very recent tissue from recent months and years. Since our practice has been to make TMAs when the included cases have long-term follow-up, we had no recent tissue cohorts to include in the study. In future studies, it will be interesting to see the change in antigenicity in newly fixed tissue compared with archived tissue. Another limitation was the use of preexisting cohorts with limited temporal overlap. We attempted to account for this potential effect in the random effects analysis and by staining all cohorts side-by-side under identical conditions, but it is possible that the differences in preparation or handling of individual cohorts could affect signal detection. In the same vein, although TMA slides were cut from arrayed blocks on the same day, coring of parent tumor blocks and assembly of the TMA block took place at different times for each cohort. Finally, an inherent limitation in the use of TMAs is the small specimen size in comparison to whole tissue sections. Though this is a strength in the sense of tissue preservation and amplification, single core samples may not be representative of a whole, heterogeneous tumor. Future investigations could incorporate side-by-side analysis of tumor specimens from multiple years or decades that are simultaneously cored.
To account for the inclusion of low- or non-expressing cases in each cohort, we used published population data to estimate the expected proportion of positive cases in each cohort. The loss of signal appears to occur in a biomarker-dependent manner. The fact that the loss is not uniform for all antigens is not surprising. This suggests that some markers may be evaluated on older cohorts without concern for aging bias, while other markers must be somehow normalized by tissue age in longitudinal studies.
While only antigenic degradation was assessed in this study, it raises suspicion regarding the degradation of other biomarkers: nucleic acid biomarkers, particularly RNA, which is unstable and thus likely to be vulnerable to degradation.15 In one study, in situ RNA detection was not measurable in tissues prior to 1993.16 DNA is thought to be most stable, but recent whole exome sequencing efforts in our group have noted increased specimen failure with age. Further understanding of age-dependent loss of biomarker information could also depend on storage conditions and other pre-analytic variables that can be optimized for signal preservation.
This investigation of tissue from the last five decades demonstrates loss of antigenicity that appears to occur within the paraffin block. While FFPE archives provide an invaluable resource for biomedical research for institutions and cooperative groups, the systematic loss of antigenicity should be considered when those archives are used for immunohistochemistry studies. Our data show that as tissue repositories continue to age, it will be important to account for the aging of the tissue in the paraffin block.
References
Simon RM, Paik S, Hayes DF . Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446–1452.
Romond EH, Perez EA, Bryant J et al. Trastuzumab plus adjuvant chemotherapy for operable Her2-positive breast cancer. N Engl J Med 2005;353:1673–1684.
Perez EA, Jenkins RB, Dueck AC et al. C-myc alterations and association with patient outcome in early-stage Her2-positive breast cancer from the North Central Cancer Treatment Group N9831 adjuvant trastuzumab trial. J Clin Oncol 2011;29:651–659.
Mass RD, Press MF, Anderson S et al. Evaluation of clinical outcomes according to Her2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer 2005;6:240–246.
Hayes DF, Allen J, Compton C et al. Breaking a vicious cycle. Sci Transl Med 2013;5:196cm196.
Camp RL, Neumeister V, Rimm DL . A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol 2008;26:5630–5637.
Bertheau P, Cazals-Hatem D, Meignin V et al. Variability of immunohistochemical reactivity on stored paraffin slides. J Clin Pathol 1998;51:370–374.
DiVito KA, Charette LA, Rimm DL et al. Long-term preservation of antigenicity on tissue microarrays. Lab Invest 2004;84:1071–1078.
Camp RL, Charette LA, Rimm DL . Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000;80:1943–1949.
Camp RL, Chung GG, Rimm DL . Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 2002;8:1323–1327.
Cheang MC, Chia SK, Voduc D et al. Ki67 index, Her2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009;101:736–750.
Wester K, Wahlund E, Sundstrom C et al. Paraffin section storage and immunohistochemistry. effects of time, temperature, fixation, and retrieval protocol with emphasis on P53 protein and Mib1 antigen. Appl Immunohistochem Mol Morphol 2000;8:61–70.
Jacobs TW, Prioleau JE, Stillman IE et al. Loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer. J Natl Cancer Inst 1996;88:1054–1059.
Blind C, Koepenik A, Pacyna-Gengelbach M et al. Antigenicity testing by immunohistochemistry after tissue oxidation. J Clin Pathol 2008;61:79–83.
Cukier HN, Pericak-Vance MA, Gilbert JR et al. Sample degradation leads to false-positive copy number variation calls in multiplex real-time polymerase chain reaction assays. Anal Biochem 2009;386:288–290.
Bordeaux JM, Cheng H, Welsh AW et al. Quantitative in situ measurement of estrogen receptor MRNA predicts response to tamoxifen. PLoS One 2012;7:e36559.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
DLR has served as a paid consultant or advisor to Cernostics, Genoptix/Novartis, Applied Cellular Diagnostics, BMS, Amgen, Optrascan, Biocept, PerkinElmer, MDAgree, and Metamark Genetics, and holds equity in Metamark Genetics. This work was supported by a gift from Gilead Sciences, the Breast Cancer Research Foundation, and grants from the NIH.
Additional information
Although archived tumor specimens provide a wealth of material for clinical investigation, such research generally relies on the assumption that the target biomarkers do not degrade with time. This paper demonstrates that there is a need for adjustment of tissue age when studying formalin-fixed paraffin embedded biospecimens. The rate of antigenicity loss is biomarker-specific and should be considered as an important variable for with studies using archived tissues.
Rights and permissions
About this article
Cite this article
Combs, S., Han, G., Mani, N. et al. Loss of antigenicity with tissue age in breast cancer. Lab Invest 96, 264–269 (2016). https://doi.org/10.1038/labinvest.2015.138
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.138
This article is cited by
-
Long term trends of breast cancer incidence according to proliferation status
BMC Cancer (2022)
-
HER2-low breast cancer shows a lower immune response compared to HER2-negative cases
Scientific Reports (2022)
-
Outcome of patients with metastatic triple negative breast cancer treated with first-line chemotherapy: a single institution retrospective analysis
Breast Cancer Research and Treatment (2022)
-
Accelerated instability testing reveals quantitative mass spectrometry overcomes specimen storage limitations associated with PD-L1 immunohistochemistry
Laboratory Investigation (2020)
-
Association between low estrogen receptor positive breast cancer and staining performance
npj Breast Cancer (2020)