Abstract
Crosstalk between the Notch signaling pathway and Caudal-related homeobox 2 (Cdx2) has important roles in the development of Barrett’s esophagus (BE). We investigated the expression and function of the Notch signaling ligand Delta-like 1 (Dll1) during the development of BE. We determined the expression levels of Dll1 and intracellular signaling molecules related to Notch signaling ((Notch1, Hairy/enhancer of split 1 (Hes1), and Atonal homolog 1 (ATOH1)) in human esophageal squamous and Barrett’s epithelium samples. Next, those expression levels in esophageal squamous cells (Het-1A) and Barrett’s esophageal cells (CP-A and BAR-T) following stimulation with either bile acids or gamma-secretase inhibitor were investigated. Finally, changes in those expression levels following transfection of a Cdx2 or Dll1 expression vector into Het-1A cells were examined. In addition, changes in those expression levels following knockdown of Cdx2 or Dll1 in CP-A cells were also examined. Dll1 was found to be upregulated and localized in the cell membrane and cytoplasm in BE. Bile acids enhanced cytoplasmic expression of Dll1 in CP-A cells, while cleaved Notch1 expression did not change, suggesting lack of a Dll1 agonistic effect on Notch signaling. Cells transfected with Cdx2 revealed significantly enhanced Dll1, while forced expression of Dll1 enhanced ATOH1, Cdx2, and MUC2 expression levels. Nevertheless, enhanced Dll1 did not induce Hes1 expression, suggesting that Dll1 may primarily function as an intracellular signaling molecule and not a Notch agonistic ligand in the canonical pathway. In addition, knockdown of Cdx2 completely abrogated any increase in Dll1 expression upon treatment with bile acids. Our results revealed a novel function of Dll1: facilitation of intestinal metaplasia in conjunction with Cdx2 expression. Furthermore, they suggest that intracellular induction of Dll1 expression in esophageal epithelial cells due to Cdx2 induction in response to bile acids has important roles in BE development.
Similar content being viewed by others
Main
Barrett’s esophagus (BE) is a premalignant condition in which normal stratified squamous epithelium of the distal esophagus is replaced by intestinal-type metaplastic columnar epithelium with goblet cells in response to chronic duodeno-gastro-esophageal reflux.1, 2 Bile acids are well known to have a critical role in the development of BE.3, 4 We previously reported a two-step mechanism involved in the development of Barrett’s epithelium, in which bile acids activate the Caudal-related homeobox 2 (Cdx2) promoter via nuclear factor kappa B (NF-κB) and stimulate production of Cdx2 protein in esophageal immature keratinocytes, resulting in the production of MUC2 (intestinal-type protein found in Barrett’s metaplasia).3
The Notch signaling pathway is essential for maintenance of cell fate such as differentiation, proliferation, and apoptosis.5 Canonical Notch signaling is activated by ligand–receptor interactions releasing the Notch intracellular domain (NICD) from proteolytic cleavage by gamma-secretase and metalloprotease. Once released, NICD translocates into the nucleus where it directly interacts with the CSL (CBF1/RBP-jκ, Su(H), Lag-1) transcriptional regulator and typically activates transcription of hairy/enhancer of split 1 (Hes1, also called HRY).6, 7 Notch target genes of the Hes family function as transcription repressors and repress atonal homolog 1 (ATOH1, also called HATH1 or Math1).8 Notch signaling controls the fate of intestinal progenitors by differentially regulating the two opposing transcriptional factors, Hes1 and ATOH1. In the esophagus, Notch signaling has also been reported to be important for esophageal epithelial homeostasis,9 as well as for the development of BE10, 11 and Barrett’s adenocarcinoma.12 In addition, we recently reported that activation of ATOH1 via Hes1 suppression in esophageal epithelial cells due to induction of Cdx2 expression in response to bile acids has important functions in induction of metaplastic changes during BE development.13 Therefore, in addition to Cdx2 expression, inactivation of Notch signaling has important roles in that development.
Among canonical Notch ligands, including Delta-like (Dll1, Dll3, and Dll4), and Serrate-like (Jagged1 and Jagged2), Dll1 is known to have an important role in the development of intestinal metaplasia, while it has also been reported that induction of Cdx2 in the intestine leads to induction of Dll1, which is involved in the mechanism of cellular differentiation from progenitor cells to goblet cells.14, 15 Given that inactivation of Notch signaling facilitates intestinal metaplasia, Dll1 contributes to goblet cell differentiation in a non-canonical manner and not as a Notch agonistic ligand. However, the detailed functions of Dll1 in the development of BE remain to be elucidated, especially regarding whether Dll1 is an important factor for driving the process of goblet cell metaplasia.
In the present study, we investigated alterations of Dll1 expression in response to bile acids in esophageal epithelial cells. Furthermore, we sought to clarify the interaction between Notch signaling and Cdx2 by focusing on the function of Dll1 in detail in the development of esophageal intestinal metaplasia, BE.
MATERIALS AND METHODS
Patients and Human Esophageal Tissues
A total of 40 patients (21 males, mean age 63.9±9.8 years) treated from August 2010 to July 2011 were enrolled in the study, including 20 with a normal esophagus and 20 with BE. The esophago-gastric junction was defined as the proximal margin of the gastric folds. Endoscopic biopsy specimens of BE were investigated in a histological manner according to the guidelines of the American College of Gastroenterology.16, 17 BE was defined histologically as columnar epithelium accompanied by goblet cell metaplasia. Biopsy samples were precisely taken from esophageal specimens and then snap-frozen in liquid nitrogen. Human tissue specimens were obtained from patients who underwent an endoscopic examination at Shimane University Hospital with a GIF-H260, H260Z, or Q260J endoscope (Olympus Medical Systems, Tokyo, Japan). All endoscopic examinations were performed by well-trained expert endoscopists (Y Tamagawa, N Ishimura, G Uno, T Yuki) and endoscopic diagnosis was established by consensus of at least three of those experts. The study protocol was prepared according to the Declaration of Helsinki and approved by the ethics committee of Shimane University Faculty of Medicine. Written informed consent was obtained from all patients.
Histopathology
Histopathological analysis was performed using tissues sectioned into 5-μm slices and stained with hematoxylin and eosin (HE) for light microscopic examinations. Biopsy specimens were immediately placed in a 10% buffered formalin solution, routinely processed, and embedded in paraffin, with seven sequential sections obtained. Two sections were stained with HE and periodic acid-Schiff (PAS) Alcian blue staining, and a histological diagnosis of BE was made when specialized columnar epithelium containing goblet cells with acid mucin was identified in the biopsy specimen by PAS Alcian blue staining.16 The remaining five sequential sections were used for immunohistochemistry examinations. When Barrett’s epithelial cells were more dominantly stained by the anti-MUC2 antibody, the case was diagnosed as BE with intestinal predominant mucin phenotype.
Immunohistochemistry
Immunolabeling was performed using formalin-fixed paraffin-embedded blocks (5-μm thick). Freshly cut sections were deparaffinized in xylene and rehydrated through sequential graded ethanol steps, then incubated with primary antibodies, followed by incubation with secondary biotinylated antibodies (DAKO, Carpenteria, CA, USA). Bound antibodies were detected using an avidin–biotin peroxidase method (ABC Elite Kit; Vector Laboratories, Burlingame, CA, USA). The sections were then counterstained with HE. The primary antibodies used in this study included goat anti-Dll1 antibody (sc-8155, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:500, while dilutions for the other primary antibodies have been previously described.13 Mounted slides were examined by light microscopy, and immunoreactivity was assessed using a three-grade system, in which 0 denoted absence of staining; 1 minimal and variable staining; and 2 obvious, uniform, and intense staining. Only specimens with grade 2 immunoreactivity were considered to be positive.
Cell Culture and Treatments
Three cell lines, Het-1A (normal human esophageal squamous epithelial cell line immortalized by viral SV40 transfection; American Type Culture Collection (ATCC), Manassas, VA, USA), CP-A (human BE cell line derived from non-dysplastic metaplasia; ATCC), and BAR-T (telomerase-immortalized Barrett’s cell line derived from endoscopic biopsy specimens of non-dysplastic specialized intestinal metaplasia taken from patients with long-segment BE), were used in this study. BAR-T cells were provided by Rhonda F Souza (The University of Texas Southwestern Medical Center, Dallas, TX, USA). The cells were maintained in monolayer cultures at 37 °C in a humidified incubator containing 5% carbon dioxide (CO2). Het-1A cells were cultured in BEBM (Lonza, Walkersville, MD, USA) medium supplemented with 100 U/ml penicillin G (Invitrogen, Carlsbad, CA, USA) and 100 μg/ml streptomycin (Invitrogen). CP-A cells were cultured in MCDB 153 medium (Omega Scientific, Tarzana, CA, USA) supplemented with 5% fetal bovine serum (FBS), 20 ng/ml recombinant human epidermal growth factor (EGF) (R&D Systems, Minneapolis, MN, USA), 0.4 μg/ml hydrocortisone (Sigma, St Louis, MO, USA), 10 nM cholera toxin, 20 mg/l adenine, 140 μg/ml bovine pituitary extract (BPE), 0.1% Insulin-Transferrin-Sodium (ITS) Selenite Supplement, and 4 mM glutamine (all from Sigma), and 100 U/ml penicillin G and 100 μg/ml streptomycin (each from Invitrogen). BAR-T cells were cultured as previously described.18 Sub-confluent cells were split, sub-cultured in plastic dishes until 70% confluent, and then serum starved 24 h before the experiments. For individual experiments, all cells were seeded into collagen I-coated wells (BD Biosciences, San Jose, CA, USA). All cell culture experiments were performed in triplicate to ensure reproducibility. After reaching approximately 70% confluence. the cells were incubated with increasing concentrations of deoxycholic acid (DCA) (Sigma; 50, 100, and 200 μM for each cell line) at neutral pH for up to 12 h or with an increasing concentration of the gamma-secretase inhibitor (GSI) N-{N-(3,5-difluorophenacetyl-L-aranyl)-S-phenylglicine}-t-butyl ester (DAPT) (Calbiochem, Darmstadt, Germany; 10, 15, and 20 μM for each cell line) for up to 24 h. Incubation time, DCA, DAPT, and concentrations were chosen according to criteria described previously.13 In preliminary experiments, cells were treated with DAPT at doses ranging from 5 to 40 μM, which showed that doses ≥30 μM decreased cell viability (preliminary data not shown). Therefore, we used DAPT for up to 20 μM in all further experiments Inhibition of Notch signaling was achieved by addition of DAPT. For experimental groups at neutral pH, the medium was titrated to the corresponding pH prior to addition of the other components, and pH was verified again thereafter. The cells were then incubated for the indicated time periods. Extraction of RNAs and proteins was performed as described below.
RNA Extraction and Real-Time PCR
Extraction of total RNA was performed as previously described.13 Prior to performing reverse transcription, all RNA samples were treated with RNase-free DNase I (Ambion, Austin, TX, USA), following the manufacturer’s instructions. For real-time RT-PCR assays, complementary DNA (cDNA) was synthesized from 0.5 μg total RNA using an AffintyScript QPCR cDNA Synthesis Kit (Stratagene, La Jolla, CA, USA), according to the manufacturer’s protocol, and the final reaction was diluted five-fold in RNase-free water. For real-time PCR, 2 μl of cDNA and primers were mixed with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in a final reaction volume of 20 μl and then assayed using a StepOnePlus Real Time PCR System (Applied Biosystems), as directed by the manufacturer. We utilized primer sequences for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2. The GAPDH subunit was used as the normalization control. Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2 mRNAs were quantitated using real-time PCR. Primers specific for Dll1 consisted of a forward primer, 5′-CTTCCCCTTCGGCTTCAC-3′, and a reverse primer, 5′-GGGTTTTCTGTTGCGAGGT-3′ (NCBI NM_005618). The primer sequences used for other genes were previously described.13 Real-time PCR products were subjected to melting curve analysis. Agarose gel electrophoresis showed the expected band. Data are expressed as the ratio of product copies/μl to copies/μl of the housekeeping gene GAPDH from the same RNA (respective complementary DNA) sample subjected to PCR.
Protein Extraction and Western Blotting Analysis
Protein extraction and western blotting analysis were performed as previously described.13 The blots were incubated overnight at 4 °C with the appropriate primary antibodies, followed by reprobing with anti-β-actin antibody as the loading control. Blots were then washed with Tris-buffered saline supplemented with 0.1% Tween 20 (TBST) and incubated for 1 h at room temperature in horseradish peroxidase-conjugated secondary antibodies (DAKO) at 1:3000. After a final wash with TBST, bound antibodies were visualized using a chemiluminescent substance (Amersham ECL Plus Western Blotting Reagents Components/Solution B; GE Healthcare) and exposed to New Amersham Hyperfilm ECL (GE Healthcare).
Immunofluorescence Cytochemistry
Het-1A, CP-A, and BAR-T cells were separately seeded at a density of 1 × 105 cells per well onto glass coverslips placed in six-well dishes and then fixed with fresh 4% paraformaldehyde and permeabilized with 0.1% Triton-X. The cells were incubated with the primary antibody against Dll1 (1:50) or isotype control for 1 h. Binding of the primary antibody was detected by rhodamine-conjugated anti-goat immunoglobulin (DAKO). The cells were nuclear counter-stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Pierce Biotechnology). Images were recorded using a fluorescence microscope (DM6000B, Leica Microsystem, Heerbrugg, Switzerland). For quantification, five fields per coverslip were randomly selected using a × 40 objective lens and were counted in a blinded manner. The percentage of Dll1-positive cells over the total number of nuclei was calculated.
Expression Plasmid and Transient Transfection
The cDNA encoding full-length mouse Dll1 (NCBI NM_007865) was amplified by PCR and cloned into a pcDNA 5/FRT/V5-His TOPO vector (Invitrogen). The plasmid clones were sequenced to confirm the constructs. Het-1A cells were separately grown in six-well dishes, and then transfected with the expression vector in each well using Lipofectamine 2000 Transfection Reagent (Invitrogen). Vector DNA without Dll1 sequences was used as a negative control. A Cdx2 expression vector was also constructed, as previously reported.3, 13, 19 Following incubation at 37 °C in a humidified incubator containing 5% CO2 for 6 h, 2 ml of normal growth medium was exchanged. Het-1A cells were collected at 48 h after transfection, and then samples were prepared and analyzed using real-time PCR, western blotting analysis, and immunofluorescence cytochemistry, as previously described. Plasmid vectors, such as Dll1 expression and Cdx2 expression vectors, were transfected at a final concentration of 2 μg/ml.
Small Interfering RNA (siRNA) Knockdown of Cdx2 or Dll1 Expression Levels
The siRNA, a specific double-stranded 20–25 nucleotide RNA sequence homologous to the target gene, were used to silence Cdx2 or Dll1 expression levels. The siRNA oligonucleotide for human Cdx2 (sc-43680) and Dll1 (sc-37200) were purchased from Santa Cruz Biotechnology. Inhibition of RNA or protein expression was assessed after transient transfection of CP-A cells with siRNA. In brief, cells were grown in six-well dishes and transiently transfected with 10 nM of pooled siRNAs using 0.4 μl/ml Lipofectamine 2000 Transfection Reagent in a total transfection volume of 2 ml of Dulbecco’s modified Eagle medium containing 10% FBS. After incubation at 37 °C in a humidified incubator containing 5% CO2 for 6 h, 2 ml of normal growth medium was exchanged. Samples were then prepared and analyzed by real-time PCR and western blotting analyses as previously described.
Statistical Analysis
All data were obtained from at least three independent experiments, and the results are expressed as the mean±s.e.m., unless otherwise indicated. The association between the immunoreactive grade and the mRNA expression levels of Dll1 and other factors associated with Notch signaling (Notch1, Hes1, and ATOH1) was evaluated by Peason’s correlation analysis. Multiple comparisons were performed with ANOVA, followed by Dunnett’s test. Differences with a P-value <0.05 were considered to be statistically significant.
RESULTS
Dll1 Expression Is Enhanced in BE
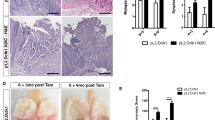
To clarify the parameters of Dll1 and other factors associated with Notch signaling (Notch1, Hes1, and ATOH1), we assessed their expression levels in human normal esophageal squamous epithelium and BE specimens using an immunohistochemistry method. The numbers and clinical characteristics of patients included in this study are shown in Table 1. No significant difference with regard to clinical characteristics was found between patients who provided normal esophagus samples and those with BE (Figure 1a). Dll1 expression was found to be localized in the cell membrane and cytoplasm in BE, while weaker and patchy staining was seen in normal esophageal specimens. Dll1 expression was observed in 80% (16 of 20) of the BE specimens with similar frequency and immunoreactivity, while that was seen in only 20% (4 of 20) of the normal esophagus specimens. In contrast, Notch1 expression was localized in the cell membrane and cytoplasm of esophageal epithelial cells in both normal esophagus and BE samples with the same immunoreactivity. Hes1 expression was clearly observed in the nucleus as well as in the cytoplasm in normal esophageal specimens, while BE specimens exhibited weaker and patchy staining. ATOH1-positive nuclear staining was observed mostly in goblet cells in BE specimens but not in normal esophageal specimens (Figure 1a). Those expression levels were confirmed by real-time PCR using endoscopic biopsy from normal human esophageal squamous epithelium and BE specimens. Consistently, Dll1 and ATOH1 mRNA expression levels were significantly greater in BE than in the normal esophageal specimens, while Hes1 mRNA expression was significantly lower in BE than in the normal esophageal specimens, with no significant difference between normal esophagus and BE for Notch1 mRNA expression levels (Figure 1b). We found a significant correlation between the immunoreactive grade and the mRNA expression levels of Dll1, Notch1, Hes1, and ATOH1 (Figure 1c). These findings indicate that Dll1 expression is enhanced in BE by inactivated Notch signaling.
Differences regarding Dll1 and other factors associated with Notch signaling (Notch1, Hes1, and ATOH1) between human normal esophageal squamous epithelium and BE specimens. (a) Immunohistochemical detection of Dll1, Notch1, Hes1, and ATOH1 in biopsy specimens taken from normal esophageal squamous epithelium and BE. Right panels show higher magnification (× 800) of boxed areas in the left panels (× 400). (b) Expression of the indicated genes was quantitated by real-time PCR in normal esophageal squamous epithelium and BE. (c) Relationship between the immunoreactive grade (0–2) and mRNA expression levels of Dll1, Notch1, Hes1, and ATOH1 was evaluated by Peason’s correlation analysis in normal esophageal squamous epithelium and BE. Data are shown as the mean±s.e.m. of at least three experiments. **P<0.01, compared with normal esophagus (n=20 for each group). NS, not significant.
Bile Acids Induce Dll1 Expression in Human Esophageal Cells
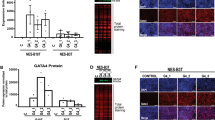
Bile acids are known to have a critical role in the pathogenesis of BE.3, 4 To determine whether they affect Dll1 expression, we investigated the effects of DCA on Dll1 expression by immunoblot analysis and real-time PCR in human esophageal squamous epithelial (Het-1A) and Barrett’s epithelial (CP-A and BAR-T) cells. Only small amounts of Dll1 mRNA expression was detectable in the quiescent Het-1A cells without any stimulation, while endogenous Dll1 mRNA expression levels was induced in all cell lines in response to DCA treatment in a concentration- and time- dependent manner, peaking at 10 h (Figures 2a and b). Dll1 protein expression was significantly increased in Het-1A, CP-A, and BAR-T cells following exposure to 200 μM DCA for 10 h (Figure 2c). Likewise, DCA upregulated Dll1 mRNA expression in these cells (Figure 2d). Immunocytochemistry findings showed that Dll1 immunoreactivity was augmented in the cell membrane and cytoplasm after administration with DCA, suggesting that bile acids induced intracellular Dll1 expression (Figures 2e and f). Furthermore, we investigated the effects of DCA on Notch signaling molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1) in Het-1A, CP-A, and BAR-T cells. Consistent with our previous report,13 DCA decreased Hes1 protein expression, while it enhanced ATOH1 protein expression, with no difference seen with regard to Notch1 and Cleaved Notch1 protein expression levels (Figure 2c). Likewise, DCA suppressed Hes1 mRNA expression in these cells, while ATOH1 mRNA expression was augmented following stimulation with DCA, with no difference in Notch1 mRNA expression (Figure 2d), indicating no Dll1 agonistic effect on Notch signaling in cells exposed to bile acids.
Exposure to DCA increased Dll1 in Het-1A, CP-A, and BAR-T cells. (a) Het-1A, CP-A, and BAR-T cells were treated with various concentrations of DCA (0–200 μM) for 10 h. Dll1 transcripts were measured by real-time PCR. (b) Time-dependent effects of DCA on Dll1 gene expression. All cell lines were harvested and subjected to total RNA isolation immediately before (0) and 8, 9, 10, 11, and 12 h after stimulation by DCA (200 μM). (c) Findings with western blotting of Dll1 and Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1) in Het-1A, CP-A, and BAR-T cells after treatment with DCA (200 μM) for 10 h. (d) mRNA expression levels of Dll1, Notch1, Hes1, and ATOH1 in all cell lines shown by quantitative real-time PCR following treatment with DCA (200 μM) for 10 h. (e) Immunofluorescence cytochemistry findings for Dll1 and (f) the percentages of Dll1-positive cells under the same conditions. Dll1 immunostaining was increased in DCA-treated Het-1A, CP-A, and BAR-T cells as compared with the control (untreated cells). Scale bars: 20 μm. Data are shown as the mean±s.e.m. of at least three experiments. *P<0.05; **P<0.01 (n=4 for each group). NS, not significant.
GSI Enhances Dll1 Expression in Human Esophageal Cells
DAPT is known as a specific GSI of the Notch signaling pathway.20, 21 To determine whether inactivation of Notch signaling by GSI affects the expression level of Dll1, we investigated the effects of DAPT on Dll1 mRNA expression in Het-1A, CP-A, and BAR-T cells and found that it increased endogenous Dll1 mRNA expression in a concentration- and time-dependent manner, peaking at 24 h (Figures 3a and b). Effective inhibition of Notch signaling was confirmed by expression of the intracellular-gamma-secretase cleaved Notch1 fragment using a specific antibody, as well as Hes1 and ATOH1 expression levels (Figures 3c and d). Furthermore, protein and mRNA expression levels of Dll1 were significantly upregulated following exposure to 20 μM DAPT in Het-1A, CP-A, and BAR-T cells. Consistent with our previous report,13 DAPT increased Cdx2 and MUC2 expression levels in these cells. These results suggest that inhibition of Notch signaling by GSI induces the expression of Cdx2 and MUC2, as well as that of Dll1.
Exposure to DAPT increased Dll1 in Het-1A, CP-A, and BAR-T cells. (a) All cell lines were treated with various concentrations of DAPT (0–20 μM) for 24 h. Dll1 transcripts were measured by real-time PCR. (b) Time-dependent effects of DAPT on Dll1 gene expression. All cell lines were harvested and subjected to total RNA isolation immediately before (0) and 12, 18, and 24 h after stimulation by DAPT (20 μM). (c) Findings with western blotting of Dll1, Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in Het-1A, CP-A, and BAR-T cells following treatment with DAPT (20 μM) for 24 h. (d) mRNA expression levels of Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2 shown by quantitative real-time PCR in Het-1A, CP-A, and BAR-T cells under the same conditions. Data are shown as the mean±s.e.m. of at least three experiments. *P<0.05; **P<0.01 (n=4 for each group). NS, not significant.
Dll1 Expression in Human Esophageal Cells Is Cdx2 Dependent
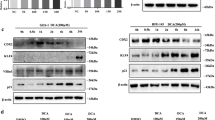
We previously demonstrated that Cdx2 overexpression regulated the Notch target genes Hes1 and ATOH1 in esophageal epithelial cells.13 To investigate whether Cdx2 overexpression influences Dll1 expression, we transfected a Cdx2 expression vector into Het-1A cells and determined the activation of Dll1 at protein and mRNA expression levels. Indeed, Dll1 expression was significantly enhanced in Cdx2-transfected cells, indicating that Dll1 expression in esophageal cells is Cdx2 dependent (Figures 4a and b). Consistent with our previous study, forced expression of Cdx2 in cells suppressed Hes1 and enhanced ATOH1 and MUC2 expression with no influence on Notch1 cleavage. Interestingly, enhanced Dll1 expression did not affect Notch1 cleavage, suggesting that Dll1 mainly functions as an intracellular signaling molecule in response to Cdx2 and not as a Notch agonistic ligand in the canonical pathway.
Transfection of Cdx2 enhanced the expression of Dll1 in Het-1A cells. (a) Findings with western blotting of Dll1, Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in Het-1A cells after transfection with an empty or Cdx2 expression vector. (b) mRNA expression levels of Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2 in Het-1A cells shown by quantitative real-time PCR under the same conditions. Data are shown as the mean±s.e.m. of at least three experiments. **P<0.01, compared with empty vector (n=4 for each group). NS, not significant.
Activation of Dll1 Increases the Expression of ATOH1, Cdx2, and MUC2 in Human Esophageal Cells
Next, we sought to investigate whether Dll1 overexpression has effects on Notch signaling and Cdx2 expression in esophageal epithelial cells. We transfected a Dll1 expression construct into Het-1A cells and determined the protein expression levels of Notch signaling molecules, as well as those of Cdx2 and MUC2. After 48 h, cells transfected with the Dll1 expression vector showed upregulated ATOH1 protein expression, while no influence was seen on Hes1 expression. Furthermore, Dll1 overexpression induced Cdx2 and MUC2 protein expression levels, with no differences observed between Dll1- and empty vector-transfected cells with regard to the protein expression levels of Notch1 and Cleaved Notch1 (Figure 5a). Consistently, cells transfected with the Dll1 expression construct showed significantly induced ATOH1, Cdx2, and MUC2 mRNA expression levels, while there was no influence on Hes1 or Notch1 mRNA expression levels in Het-1A cells (Figure 5b). To confirm the localization of Dll1 expression in cells transfected with the Dll1 expression vector, we performed immunocytochemistry for Dll1 using Het-1A cells. Dll1 protein expression was augmented, as shown by positive cell membrane and cytoplasm staining, after transfection of Dll1 in the same manner as cells exposed to DCA (Figures 2e, f, 5c, and d).
Transfection of Dll1 enhanced the expression of ATOH1, Cdx2, and MUC2 in Het-1A cells. (a) Findings with western blotting of Dll1, Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in Het-1A cells after transfection of an empty or Dll1 expression vector. (b) mRNA expression of Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2 in Het-1A cells shown by quantitative real-time PCR under the same conditions. (c) Immunofluorescence cytochemistry of Dll1 and (d) the percentages of Dll1-positive cells under the same conditions. Dll1 immunostaining was increased in Dll1-transfected Het-1A cells as compared with the control (empty vector-transfected Het-1A cells). Scale bars: 20 μm. Data are shown as the mean±s.e.m. of at least three experiments. **P<0.01, compared with empty vector (n=4 for each group). NS, not significant.
Dll1 Induction by Bile Acids is Cdx2 Dependent in Human Barrett’s Cells
To determine whether Dll1 induction by bile acids occurs via Cdx2 activation, we employed the siRNA approach to inhibit endogenous Cdx2 in CP-A cells. In Cdx2 siRNA-transfected samples, Cdx2 protein expression were significantly blocked (Figure 6a). Furthermore, the protein expression levels of Dll1, ATOH1, and MUC2 were significantly decreased, whereas Hes1 protein expression was upregulated in cells transfected with Cdx2-specific siRNA as compared with the control nonspecific siRNA-transfected cells following bile acid treatment. In contrast, the protein expression levels of Notch1 and Cleaved Notch1 were not affected by treatment with the siRNA targeted to Cdx2 (Figure 6a). Consistently, knockdown of Cdx2 significantly downregulated the mRNA expression levels of Dll1, ATOH1, and MUC2, whereas Hes1 mRNA expression was increased in cells treated and not treated with bile acids, and there was no influence on Notch1 mRNA expression (Figure 6b). These data suggest that Dll1 induction by bile acids is Cdx2 dependent in Barrett’s epithelial cells.
Dll1 induction by bile acid is Cdx2 dependent in CP-A cells. (a) Findings with western blotting of Dll1, Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in CP-A cells after transfection of either control nonspecific siRNA or Cdx2-specific siRNA with or without DCA treatment (200 μM) for 10 h. (b) mRNA expression of Dll1, Notch1, Hes1, ATOH1, and MUC2 in CP-A cells shown by quantitative real-time PCR under the same conditions. Data are expressed as the n-fold increase in transcripts in DCA-stimulated samples over that in the vehicle-treated samples. Results are shown as the mean±s.e.m. of at least three experiments. **P<0.01, compared with control siRNA-transfected samples with DCA treatment (n=4 for each group). NS, not significant.
Knockdown of Dll1 Suppresses the Expression of ATOH1, Cdx2, and MUC2 in Human Barrett’s Cells
Finally, we sought to investigate whether knockdown of Dll1 by treatment with Dll1 siRNA affects the expression of ATOH1, Cdx2, and MUC in CP-A. In Dll1 siRNA-transfected samples, both Dll1 protein and mRNA expression levels were significantly blocked (Figures 7a and b). In addition, the protein expression levels of ATOH1, Cdx2, and MUC2 were significantly suppressed, with no differences observed between Dll-1-specific siRNA-transfected and the control nonspecific siRNA transfected cells with regard to the protein expression levels of Notch1, Cleaved Notch1, and Hes1 (Figure 7a). Consistently, knockdown of Dll1 significantly downregulated the mRNA expression levels of ATOH1, Cdx2, and MUC2, while there was no influence on Hes1 or Notch1 mRNA expression levels in CP-A cells (Figure 7b).
Knockdown of Dll1 suppressed the expression of ATOH1, Cdx2, and MUC2 in CP-A cells. (a) Findings with western blotting of Dll1, Notch molecules (Notch1, Cleaved Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in CP-A cells after transfection of either control nonspecific siRNA or Dll1-specific siRNA. (b) mRNA expression of Dll1, Notch1, Hes1, ATOH1, Cdx2, and MUC2 in CP-A cells shown by quantitative real-time PCR under the same conditions. Data are shown as the mean±s.e.m. of at least three experiments. **P<0.01, compared with control siRNA-transfected samples (n=4 for each group). NS, not significant.
DISCUSSION
The major aim of this study was to determine whether Dll1 expression is related to the development of BE. Although Dll1 has been reported to be enhanced in intestinal metaplasia in the small and large intestines,14, 15 little is known about the molecular mechanism of Dll1 in the development of BE. Our findings showed that Dll1 expression was upregulated and localized in the cell membrane and cytoplasm in BE specimens but not in those of esophageal squamous epithelium. In addition, Dll1 was induced by bile acids in conjunction with Cdx2 expression in esophageal epithelial cells. These results are the first to show that Dll1 is an important molecular mediator induced by bile acids in BE development and its function in that development is discussed in greater detail below.
In addition to their well-characterized role of activating Notch signaling via cell-to-cell interactions, Notch ligands can also affect Notch signaling through interactions with Notch within the same cell.22, 23 Intracellular interaction between Notch ligands and Notch was shown to inhibit Notch signaling.22, 23, 24, 25 Therefore, Dll1, a Notch ligand, may function as both a Notch signaling agonist and antagonist in the development of BE. If Dll1 functions as a canonical Notch ligand, then the Notch receptor is cleaved by gamma-secretase and metalloprotease, while the downstream transcription factor Hes1 is induced and ATOH1 expression is suppressed. However, we previously showed that inhibition of Notch signaling in esophageal epithelial cells triggered trans-differentiation to intestinal goblet-type columnar epithelial cells.13 Moreover, suppressed Hes1 and forced ATOH1 expression by inhibition of Notch signaling with GSI is known to promote MUC2 via Cdx2 expression in both esophageal epithelial and esophageal adenocarcinoma cell lines. Consistent with our previous study, the present findings showed inactivation of Notch signaling, indicating suppressed Hes1 and forced ATOH1 expression, in the human Barrett’s cell lines CP-A and BAR-T. Therefore, it is suggested that Dll1 has a role in facilitating goblet cell metaplasia, which is distinct from its function as a Notch agonistic ligand in the canonical pathway during the development of BE.
Bile acids, particularly unconjugated bile acids such as DCA, are a component of gastro-duodenal reflux that have been strongly linked to BE development.26, 27, 28 In previous studies, DCA showed the strongest effect on Cdx2 transcription of all bile acids.29, 30, 31 In our earlier reports, we proposed that bile acids suppress the expression of Hes1 via Cdx2 activation in esophageal epithelial cells, with resulting activation of ATOH1 and MUC2.3, 13 In the present study, Dll1 expression was upregulated in Het-1A, CP-A and BAR-T cells by exposure to DCA. In addition, DCA inhibited the downstream target of Notch signaling Hes1 without affecting Notch 1 cleavage. Therefore, DCA may induce Dll1 expression, while induced Dll1 does not act as a Notch ligand in the canonical pathway. Indeed, our immunocytochemistry findings showed that DCA induced intracellular Dll1 protein in all the cell lines. We also found that knockdown of Cdx2 by treatment with Cdx2 siRNA completely abrogated any increase in Dll1 expression upon treatment with DCA, suggesting that Dll1 induction by bile acids in Barrett’s epithelial cells is a Cdx2-dependent process.
Notch signaling has an essential role in cell fate in intestinal epithelium, and when blocked, proliferative epithelial cells instantly change to goblet cells.32, 33, 34 Interestingly, inactivation of Notch signaling by GSI is associated with increased levels of Dll1. Moreover, GSI may induce Cdx2 and MUC2 expression by overexpression of ATOH1 via Hes1 suppression. Consistent with our findings, a previous study showed that GSI suppressed Hes1 and enhanced ATOH1 expression in human colonic epithelial cells, with resulting activation of Dll1 and MUC2, indicating that Dll1 has an important role in goblet cell differentiation in human colonic tissue.14 Collectively, inactivation of Notch signaling by either bile acids or GSI affects Dll1 expression associated with Cdx2 expression.
Although Cdx2 is known to have important roles in the development of BE,3, 35, 36 the inter-regulation mechanism between Cdx2 and Dll1 remains to be elucidated in detail. Recently, Grainger et al15 demonstrated that the Dll1 promoter was occupied by Cdx response elements, while Cdx2-dependent Dll1 expression played important roles in both somitogenesis and goblet cell differentiation, suggesting that Dll1 operates downstream of Cdx2, possibly via a direct regulating interaction. Indeed, forced expression of Cdx2 in Het-1A cells induced the expression of Dll1 and MUC2, whereas it suppressed that of Hes1. In addition, a previous report noted that Dll1 was a direct target of Hes1 in mice and also showed that Hes1 suppression caused by blocking Notch signaling induces Dll1 expression in neural progenitors.37 Together, these results strongly suggest that Dll1 mainly functions as an intracellular signaling molecule and is regulated by Cdx2 and Hes1 expression. When Het-1A cells were transfected with the Dll1 expression vector, forced expression of Dll1 in those cells induced the expression of ATOH1, Cdx2, and MUC2, while no influence on upstream Notch signaling (Notch1, Cleaved Notch1, and Hes1) was noted. Consistent with our present findings, Akiyama et al14 reported that Dll1 was directly involved in transcriptional regulation of the ATOH1 gene in Notch-inactivated human intestinal epithelium. Furthermore, knockdown of Dll1 by treatment with Dll1 siRNA significantly attenuated the expression of ATOH1, Cdx2, and MUC2 in CP-A cells. Therefore, it is considered that forced expression of Dll1 increases Cdx2 expression via ATOH1 activation in esophageal epithelial cells, while crosstalk between Dll1 and Cdx2 contributes to induce goblet cell formation in the development of BE. Taken together with our previous findings,13 our present results show a novel function of Dll1 to promote the development of intestinal metaplasia induced by bile acids in a Cdx2-dependent manner (Figure 8). Recent findings have consistently indicated that the intracellular domains of Notch ligands function as transcriptional co-activators24 and facilitate transcription of various genes,23, 24, 25 including ATOH1, Cdx2, and MUC2. Nonetheless, further research is required for elucidation of the precise role of Dll1 as a regulator of intracellular transcription in BE development.
Proposed molecular model of Dll1 in induction of metaplastic changes during development of BE. Bile acids directly stimulate Cdx2 expression. Overexpression of Cdx2 directly induces MUC2, whereas that of ATOH1 indirectly induces MUC2 via Cdx2 expression. Thereafter, overexpression of Cdx2 induces ATOH1 and Dll1 expression levels via Hes1 suppression by blocking Notch signaling. Overexpression of Dll1 indirectly induces MUC2 expression via ATOH1 and Cdx2 expression levels.
In conclusion, our results revealed that facilitation of intestinal metaplasia development is a novel function of Dll1 in conjunction with Cdx2 expression. In addition, they suggest that intracellular induction of Dll1 in esophageal epithelial cells caused by Cdx2 induction in response to bile acids has important roles in the development of BE.
References
Spechler SJ . Barrett’s esophagus and risk of esophageal cancer: a clinical review. JAMA 2013;310:627–636.
Spechler SJ, Souza RF . Barrett’s esophagus. N Engl J Med 2014;371:836–845.
Kazumori H, Ishihara S, Rumi MA et al. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett’s epithelium. Gut 2006;55:16–25.
Huo X, Zhang HY, Zhang XI et al. Acid and bile salt-induced CDX2 expression differs in esophageal squamous cells from patients with and without Barrett's esophagus. Gastroenterology 2010;139:194–203.
Artavanis-Tsakonas S, Rand MD, Lake RJ . Notch Signaling: cell fate control and signal integration in development. Science 1999;284:770–776.
Gordon WR, Arnett KL, Blacklow SC . The molecular logic of Notch signaling - a structural and biochemical perspective. J Cell Sci 2008;121:3109–3119.
Feder JN, Li L, Jan LY et al. Genomic cloning and chromosomal localization of HRY, the human homolog to the Drosophila segmentation gene, hairy. Genomics 1994;20:56–61.
Ben-Arie N, McCall AE, Berkman S et al. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet 1996;5:1207–1216.
Ohashi S, Natsuizaka M, Yashiro-Ohtani Y et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 2010;139:2113–2123.
Morrow DJ, Avissar NE, Toia L et al. Pathogenesis of Barrett’s esophagus: bile acids inhibit the Notch signaling pathway with induction of CDX2 gene expression in human esophageal cells. Surgery 2009;146:714–722.
Menke V, van Es JH, de Lau W et al. Conversion of metaplastic Barrett’s epithelium into post-mitotic goblet cells by γ-secretase inhibition. Dis Model Mech 2010;3:104–110.
Mendelson J, Song S, Li Y et al. Dysfunctional transforming growth factor-β signaling with constitutively active notch signaling in Barrett's esophageal Adenocarcinoma. Cancer 2011;117:3691–3702.
Tamagawa Y, Ishimura N, Uno G et al. Notch signaling pathway and Cdx2 expression in the development of Barrett’s esophagus. Lab Invest 2012;92:896–909.
Akiyama J, Okamoto R, Iwasaki M et al. Delta-like 1 expression promotes goblet cell differentiation in Notch-inactivated human colonic epithelial cells. Biochem Biophys Res Commun 2010;393:662–667.
Grainger S, Lam J, Savory JG et al. Cdx regulates Dll1 in multiple lineages. Dev Biol 2012;361:1–11.
Sampliner RE . Practice guidelines on the diagnosis, surveillance, and therapy of Barrett’s esophagus. The Practice Parameter Committee of the American College of Gastroenterology. Am J Gastroenterol 1998;93:1028–1032.
Wang KK, Sampliner RE . Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett’s esophagus. Am J Gastroenterol 2008;103:788–797.
Wang DH, Tiwari A, Kim ME et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. J Clin Invest 2014;124:3767–3780.
Kazumori H, Ishihara S, Takahashi Y et al. Roles of Kruppel-like factor 4 in oesophageal epithelial cells in Barrett’s epithelium development. Gut 2011;60:608–617.
Geling A, Steiner H, Willem M et al. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep 2002;3:688–694.
Ben-Arie N, Bellen HU, Armstrong DL et al. Math1 is essential for genesis of cerebellar granule neurons. Nature 1997;390:169–172.
Ikeuchi T, Sisodia SS . The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “gamma-secretase” cleavage. J Biol Chem 2003;278:7751–7754.
LaVoie MJ, Selkoe DJ . The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem 2003;278:34427–34437.
Hiratochi M, Nagase H, Kuramochi Y et al. The Delta intracellular domain mediates TGF-beta/Activin signaling through binding to Smads and has an important bi-directional function in the Notch-Delta signaling pathway. Nucleic Acids Res 2007;35:912–922.
Kolev V, Kacer D, Trifonova R et al. The intracellular domain of Notch ligand Delta1 induces cell growth arrest. FEBS Lett 2005;579:5798–5802.
Zhang F, Altorki NK, Wu YC et al. Duodenal reflux induces cyclooxygenase-2 in the esophageal mucosa of rats: evidence for involvement of bile acids. Gastroenterology 2001;121:1391–1399.
Tselepis C, Morris CD, Wakelin D et al. Upregulation of the oncogene c-myc in Barrett's adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut 2003;52:174–180.
Quante M, Bhagat G, Abrams JA et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell 2012;21:36–51.
Hu Y, Williams VA, Gellersen O et al. The pathogenesis of Barrett's esophagus: secondary bile acids upregulate intestinal differentiation factor CDX2 expression in esophageal cells. J Gastrointest Surg 2007;11:827–834.
Hu Y, Jones C, Gellersen O et al. Pathogenesis of Barrett esophagus: deoxycholic acid up-regulates goblet-specific gene MUC2 in concert with CDX2 in human esophageal cells. Arch Surg 2007;142:540–544.
Avissar NE, Toia L, Hu Y et al. Bile acid alone, or in combination with acid, induces CDX2 expression through activation of the epidermal growth factor receptor (EGFR). J Gastrointest Surg 2009;13:212–222.
Leow CC, Romero MS, Ross S et al. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res 2004;64:6050–6057.
Yang Q, Bermingham NA, Finegold MJ et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 2001;294:2155–2156.
van Es JH, van Gijn ME, Riccio O et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 2005;435:959–963.
Phillips RW, Frierson HF Jr, Moskaluk CA . Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. Am J Surg Pathol 2003;27:1442–1447.
Liu T, Zhang X, So CK et al. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis 2007;28:488–496.
Shimojo H, Ohtsuka T, Kageyama R . Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 2008;58:52–64.
Acknowledgements
We greatly appreciate the excellent technical support provided by Ms Keiko Masuzaki. We also thank Rhonda F Souza for kindly providing the BAR-T cell line. This work was supported in part by Support for Young Staff of Shimane University 2012 (to Yuji Tamagawa), grants-in-aid for Scientific Research (23590917 and 26460937 to Norihisa Ishimura), and a grant-in-aid for Young Scientists (24790698 to Yuji Tamagawa) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Crosstalk between the notch signaling pathway and the transcription factor Cdx2 plays an important role in the development of Barrett's esophagus (BE). This study reveals a novel function for the notch ligand delta-like 1 (Dll1) in intestinal metaplasia: Dll1 primarily functions as an intracellular signaling molecule and not a notch agonistic ligand in the canonical pathway.
Rights and permissions
About this article
Cite this article
Tamagawa, Y., Ishimura, N., Uno, G. et al. Bile acids induce Delta-like 1 expression via Cdx2-dependent pathway in the development of Barrett’s esophagus. Lab Invest 96, 325–337 (2016). https://doi.org/10.1038/labinvest.2015.137
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.137
This article is cited by
-
Origins of Metaplasia in Barrett’s Esophagus: Is this an Esophageal Stem or Progenitor Cell Disease?
Digestive Diseases and Sciences (2018)
-
Gastroesophageal reflux and Barrett’s esophagus: a pathway to esophageal adenocarcinoma
Updates in Surgery (2018)
-
Reflux esophagitis and its role in the pathogenesis of Barrett’s metaplasia
Journal of Gastroenterology (2017)