Abstract
MALDI Imaging mass spectrometry has entered the field of tissue-based research by providing unique advantages for analyzing tissue specimen in an unprecedented detail. A broad spectrum of analytes ranging from proteins, peptides, protein modification over small molecules, drugs and their metabolites as well as pharmaceutical components, endogenous cell metabolites, lipids, and other analytes are made accessible by this in situ technique in tissue. Some of them were even not accessible in tissues within the histological context before. Thereby, the great advantage of MALDI Imaging is the correlation of molecular information with traditional histology by keeping the spatial localization information of the analytes after mass spectrometric measurement. This method is label-free and allows multiplex analysis of hundreds to thousands of molecules in the very same tissue section simultaneously. Imaging mass spectrometry brings a new quality of molecular data and links the expert discipline of pathology and deep molecular mass spectrometric analysis to tissue-based research. This review will focus on state-of-the-art of MALDI Imaging mass spectrometry, its recent applications by analyzing tissue specimen and the contributions in understanding the biology of disease as well as its perspectives for pathology research and practice.
Similar content being viewed by others
Main
Tissues are the primary manifestation of a very broad spectrum of diseases and are therefore one of the main pillars in clinical research and diagnostics. The investigation of morphological and molecular tissue features is the basis of generating knowledge about clinicopathological entities, diagnostics, prediction, and prognosis. Of great importance is the increased understanding of the molecular basis of diseases that provide important insights into the mechanism of diseases and augment observations of tissue morphology.1 Among the many advanced technologies used for tissue-based research, mass spectrometry might have an important role due to the unique advantages of high sensitivity, wide range of molecules (Table 1), molecular specificity, and the flexibility to address many varied analytes on a single platform.1 It allows the unlabeled and multiplex analysis of a broad variety of molecular classes in biological samples. Recently, Imaging mass spectrometry has matured, and this process is still going on, to a degree that now enables the translation of molecular information to tissue morphology. A combination of the analytical capabilities of mass spectrometry with microscopic information helps to investigate and understand molecular processes happening in specific cell types within a tissue. Among the several mass spectrometry ionization techniques that can be used to directly analyze tissues, Marix-Assisted Laser Desorption Ionization (MALDI) has led the way in the development of biological and clinical applications for Imaging mass spectrometry and is therein one of the most commonly used techniques.2, 3, 4 MALDI Imaging has already entered disciplines beside medicine, microbiology as a standard technology for classification or plant pathology for investigating spatially resolved molecular information. This technology provides a broad range of features, which should be recognized by medicine- or pathology-related investigations. In this current review, we discuss the state-of-the-art for MALDI Imaging mass spectrometry, its recent applications to analyze tissue specimen, the contributions in understanding pathophysiological processes and medicine, and sum up where the MALDI Imaging journey is heading.
MALDI IMAGING MASS SPECTROMETRY—PRINCIPLE
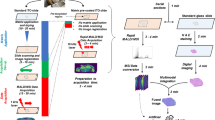
The basic principle of this method is excellently reviewed in detail previously.2 The principal workflow of a MALDI Imaging experiment is depicted in Figure 1. Materials are all types of tissue sections. These sections are covered with a matrix for extracting molecules from the tissue specimen into the matrix and aids desorption/ionization for the further analysis in the mass spectrometer. For the ionization process, the laser shoots only in the matrix layer, while the underlying tissue remains intact and thus allowing histological tissue examination after the measurement in the very same tissue section.5 The matrix absorbs the laser energy and transfers the analytes to the gas phase, promoting ionization in the process.6, 7 By selecting different matrices and depending on the technology used (MALDI-TOF or high resolution mass spectrometry as, eg, MALDI-FT-ICR), the analyte classes can be chosen.2 In the mass spectrometer, the tissue specimen are then raster-scanned (with a spatial resolution ranging from approximately 200 μm down to 20 μm), generating a mass spectrum for each measuring spot.2 Recently, a method was developed for combining a spatial resolution in the low micrometer range and high mass accuracy for the analysis of biological samples.8 Phospholipids, neuropeptides, and drug compounds were imaged with accurate mass at a pixel size between 5 and 10 μm.8, 9, 10 The molecular range that could be analyzed depends on the technique which is chosen for imaging mass spectrometry. With SIMS (secondary ion mass spectrometry), eg, detectable ions are typically limited to a narrow mass range of only a few hundred Daltons; however, cluster ion sources (eg, C60+ and Bi3+) have effectively extended this limit to ∼2 kDa.11 High-resolution imaging mass spectrometry, as MALDI-FT-ICR, has its strength more in the field of low molecular weight compounds, while MALDI-TOF is used for analyzing peptides and small proteins up to 25 kDa.12, 13, 14 Recently, several publications demonstrated that higher mass proteins could also be detected and imaged. A new detector enabled proteins up to 70 kDa to be imaged, and proteins up to 110 kDa to be detected, directly from tissue, indicating new directions by which the mass range amenable to MALDI Imaging MS and MALDI profiling MS may be extended.15 The use of the matrix ferulic acid remarkably increased signal acquisition in the mass range of 20k to 150k.16
Principle of MALDI Imaging mass spectrometry. A conventional tissue section is coated with matrix, which extracts molecules from the tissue. Afterwards, the sample is measured in a raster process in the mass spectrometer resulting in spatially resolved mass spectra, while the UV-laser only hits the matrix crystals by unaffecting the tissue section. Subsequent to the MALDI measurement, a histological staining is carried out, therefore allowing a histology-directed analysis of the mass spectra.
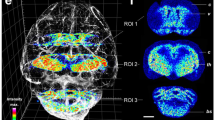
Subsequent to MALDI Imaging measurement, the very same tissue section can be stained, eg, by H&E or immunohistochemistry and coregistered to the mass spectrometric analysis. The mass signals (m/z-species) detected are then visualized as color intensity maps assigning molecular patterns to cell types. These color signals allow the detection of patterns, which represent the distribution in tissue of the molecules of interest.2 This visualization is very close to what can be seen in immunohistochemistry but in contrast with the advantage that multiple molecules can be examined by one measurement. The software solutions not only allow the visualization of individual molecules or groups of molecules but is also possible to generate, eg, cell type-specific molecular signatures (Figure 2). This, in principle, works as for tissue microdisection.17, 18 In contrast to tissue microdisection, in MALDI Imaging microdisection is performed ‘virtually’ by defining regions-of-interest (ROIs) and extracting mass spectrometric profiles within these ROIs by software. This has the great advantage, in that all tissue parts can be examined at once but separately.
Virtual microdisection. (a) A breast cancer tissue composed of cancer cells and tumor stroma is depicted (H&E) together with a MALDI Imaging average mass spectrum over the whole tumor section. (b) The green-colored mass represents a molecule, which is localized specifically in the tumor stroma, while the red colored mass is present in cancer cells. (c) To generate specific mass spectra for tumor stroma or cancer cells, virtual microdisection is performed. (d) An overlay of tumor stroma and cancer cell-specific mass spectra is depicted. Asterisk highlighting masses that are differentially expressed in tumor stroma or cancer cells demonstrating the advances of the MALDI Imaging approach in comparison to the liquid-based techniques that are not able to separate between different tissue compounds.
The identification of molecules out of mass spectra was and is challenging. In the last few years, several databases became available that now make the identification of molecules easier. In the field of cell metabolites, eg, the databases such as MetLin or Kegg can be used. Especially the results from high resolution mass spectrometers allow the identification of the peaks via those databases because they provide a high mass accuracy. This was also confirmed by a validation approach in which LC-(MS/MS) was used to validate m/z-species from high-resolution mass spectrometry which were identified previously by database alignment.19 But also in the field of protein imaging several databases are available for the identification of molecules.20, 21
UNIQUE CAPABILITIES IN TISSUE IMAGING
Proteins
When MALDI Imaging mass spectrometry entered the field of tissue-based research, the technology was mainly applied to analyze the spatial distribution of proteins.4 The imaging of proteins can deliver information about the distribution of single proteins, protein isoforms or modifications as well as posttranslational modifications, cleavage, or degradation. The information that can be achieved is comparable to immunohistochemistry but with the advantage of measuring hundreds of proteomic analytes simultaneously, which again is comparable to liquid-based proteomics but without losing spatial information (Table 2). Furthermore, eg, histone modifcations have only limited accessibility for immunohistochemical approaches; rather their investigation is a domain of a mass spectrometric approach.22 As a discovering tool for proteomic signatures and biomarkers, MALDI Imaging is implemented in a steadily increasing number of publications. Most of these studies are carried out on the use of fresh frozen tissues, but also the investigation of formalin-fixed and paraffin-embedded (FFPE) tissues is in common.2 This is an entry for investigating archive material in pathology. Furthermore, the generation of tissue microarrays (TMA) from the FFPE tissues allows high-throughput screening of tissues to define molecular patterns. This strategy was also successfully applied for TMAs of pancreatic cancer, lung tumors, gastric cancer, esophageal cancer, prostate cancer, renal cell cancer, or bladder cancer.23, 24, 25, 26, 27, 28, 29 It should be mentioned that with MALDI Imaging it is even possible to analyze the smallest biopsies from, eg, a pretherapeutic state of patients that afterwards could be correlated with therapy response to generate predictive markers.30
Lipids
Lipids are major structural components of all cell membranes and have a crucial role in the cell metabolism of all organisms.8 They represent a molecular class that had only limited accessibility for in situ analysis before. In particular, phospholipid derivatives have an important role in signal transduction and are thus of high relevance for numerous pathological processes.8, 31, 32 Changes in lipid metabolism are major factors in cancer.8, 33, 34 Imaging of lipids is now being intensively investigated using mass spectrometric imaging.8, 35, 36, 37, 38 These studies are generally performed on fresh frozen tissue, as other preservation processes deplete the lipid population. In tissue-based research, the first studies have been carried out in calf eyes to localize phospholipids. 39, 40, 41 Along the same lines, imaging of phospholipids and cholesterol after direct analysis was first done using TOF-SIMS in mice, which allowed for the mapping of these molecules in a tissue section then in MALDI Imaging.42, 43 MALDI Imaging was also used to generate lipid maps of brain and liver tissue from primates, mice, and rats or invertebrate.14, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57 The lipid metabolism was investigated in a study of 34 human colorectal cancer tissues to identify a cancer-specific phospholipid signature.58
Cell Metabolism
The analysis of endogenous metabolite profiles in tissues can lead to a deeper understanding of disease-related mechanisms. The most frequently used strategy for researching the metabolome is the use of mass spectrometry techniques, such as LC-MS or GC-MS (gas chromatography-mass spectrometry). Here too, the use of these techniques leads to loss of information relating to the localization of the analytes in the tissue. In this case, use of the MALDI Imaging technique led to a new quality of results owing to the incorporation of spatially resolved assignment of analytes. As this technique was developed further, it soon became possible to detect a great variety of components of the cell metabolism. ln a model of cerebral perfusion disorders, >30 metabolites, as well as their specific individual distribution were identified in a rat’s brain.59 A further study on the same disease model reported spatiotemporal changes in energy metabolism in connection with focal ischemia.60 In metastatic colon cancer cells in a mouse liver, the coordination between energy metabolism and cell-cycle progression of cancer cells in a xenograft model could be demonstrated. In the tumor-bearing livers, it was demonstrated that G1-phase cells were more active in glycolysis than those in S and G2–M phases.61
Drugs and Drug-related Metabolites
Former drug distribution studies were mainly carried out by the use of autoradiography. In this technique, the compound of interest had to be labeled with radioactive chemicals. In each experiment, only one compound could be analyzed at once. MALDI Imaging now brings some great advantages in this field. As a label-free and multiplex technology, it is now possible to investigate a drug and its metabolites in one and the same measurement and study their distribution in relation to tissue morphology. Additionally, MALDI Drug Imaging is also quantitative that allows studying, eg, the pharmacokinetic of drugs and each metabolite directly in tissues.62, 63, 64 Also, the field of pharmaceutical studies have markedly benefited from MALDI Drug Imaging as a method to study the tissue distribution of drugs and their metabolites throughout multiple organs.65, 66, 67, 68, 69, 70 In general, in these studies, animals are dosed with drugs, later tumors, organs, or even the whole animal body are sliced and drugs and their metabolites are analyzed.71, 72, 73, 74 In the drug discovery process in the pharmaceutical industry, essential information can be gleamed from knowing the uptake of a drug to its target as well as its metabolic pathway processes and the sites at which these are occurring within the body following administration.75 These absorption, distribution, metabolism, and excretion data are required to fully understand the efficacy, safety, and thus viability of a compound, and the earlier this is understood during the drug discovery process the increased potential exists for the ability to adapt to potential problems and subsequent time and cost savings.75 Also in human lung carcinoma tissue, personalized drug characterization with localizations was reported.76 Several MALDI Imaging reviews about drug analyses have been published so far.8, 65, 77, 78, 79, 80 An overview of drugs and their metabolites studied by MALDI Imaging so far can be found in the reviews by Prideaux and Stoeckli65 and Sun and Walch.77
Obtaining Pathways Directly from Tissue
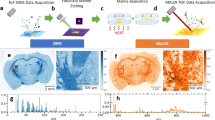
The rapid development of the MALDI Imaging technology has the potential to actively transform the field of modern tissue-based research to examine ‘omics’ information in situ in an unprecedented detail. The integration of cell-specific data without reducing, eg, tumor cell-specific effects by mixing them up with stromal cells as it is unavoidable in technologies that are based on tissue homogenates, will allow the generation of deeper molecular multi-layer information than it was possible until now with liquid-based techniques. To get a more holistic understanding of diseases and pathophysiology, it is a must to move from the single ‘omic’ information to the integrated use of multiple ‘omics’ information and to integrate this information into systems medicine tools as it already in the pipline for liquid-based techniques. Recent developments in the MALDI Imaging technology regarding higher mass accuracy (MALDI-FT-ICR) and also spatial resolution now allow the identification of molecules directly from the mass spectrometric measurement and the alignment to existing molecular databases. This information, in turn, could then be integrated into pathway analysis, thus enabling the identification of pathways directly from tissue and individual cell types.19 Such approaches might be of great interest, eg, with regards to a deeper understanding of the mechanisms of action of drugs and an organism’s individual reactions to active substances. Combining, eg, information about the distribution of the drug and its metabolites with its underlying cell metabolic and proteomic, lipidomic, etc., information will enable us to gain deep insides into the mechanisms of action. Another example also comes out of the field of pharmacological research; the investigation of drug distribution is of high interest. With the MALDI Imaging technology, it is possible, eg, to study the distribution of a drug determined by MALDI Imaging and coregistered to the immunohistochemical-stained section with, eg, CD31 to examine the drug distribution in relation to vascularization as novel approach of MALDI drug imaging, immunohistochemistry, and digital image analysis for drug distribution studies in tissues (Figure 3).81
MALDI Imaging of a Topoisomerase-2 inhibitor in a colon cancer mouse model. The MALDI Imaging measurement was coregistered to CD31 immunohistochemical-stained tumor section allowing the investigation of drug distribution in relation to vascularization (according to Buck et al 106).
SELECTED CLINICO-PATHOLOGICAL APPLICATIONS
Therapy Response Prediction and Prognosis
Molecular profiles might be of particular interest because the proteins associated with these mass signatures represent possible markers that could be further potential therapeutical targets or enable outcome predictions. A mass spectrometric signature based on seven proteins serves as a new independent indicator of unfavorable overall survival after surgical resection of gastric cancer.82 One of the first biomarker discovery studies using MALDI Imaging mass spectrometry was carried out in a patient collective in which proteomic markers in tumor tissue were identified that were associated with the response to a neoadjuvant therapy with Paclitaxel in breast cancer.83 In another study, specific biomarkers were identified in diseased lymph node tissue to distinguish between Hodgkins lymphoma and lymphadenitis.84 But it is also possible to detect protein modifications as recently demonstrated for histone modifications in hepatocellular carcinomas.22 In a recently published study, the use of MALDI Imaging facilitated the detection of previously unrecognized defects in the mitochondrial respiratory chain, which lead to individual patient response to cisplatin-based chemotherapy in advanced adenocarcinoma of the esophagus.30
Tissue-based Disease Classification
MALDI Imaging further on can be used to differentiate among tumors of varying subtype, tumor stage, or degree of metastasis. The correct identification of the tumor origin is crucial for a personalized, individually tailored treatment regimen. But in a significant number of metastasized cancers, the primary tumor cannot be identified and thus they are diagnosed as cancer of unknown primary.85, 86 Clinical diagnosis typically relies on histological and often extensive immunohistochemical analyses of tumor tissues.87 With MALDI Imaging, molecular tissue classification can be generated with a set of molecular parameters that allow differentiation among different tissues. Recently, proteomic profiles have been published that could differentiate between six adenocarcinoma entities.88 lt is also possible to detect protein signatures reflecting molecular markers, for example, predictive ones, which are already known. Two studies have accordingly described a protein profile that predicts HER2 receptor status in carcinomas of the breast and stomach by means of MALDI Imaging.89, 90 Thereby, it was not only possible to discriminate the tissues according to their HER2 status as it is already possible by immunohistochemistry, the MALDI Imaging data may provide new insights into further proteins that could be also used as predictive signature panels. A recently published study demonstrated the correct classification of breast and pancreatic cancer metastases according to MALDI Imaging profiles.91 Also, the differentiation of Spitz nevi from Spitzoid malignant melanomas has been demonstrated.92 Of course, MALDI Imaging is not limited only to be applied in cancerous tissue. MALDI Imaging was used to generate proteomic patterns to differentiate ulcerative colitis from Crohn’s colitis.93
Intertumoral and Intratumoral Heterogeneity
The reflection of molecular information in context to morphology is of special importance in tissue-based research, particularly for investigating intratumoral heterogeneity. Intratumor heterogeneity is an important factor influencing the evolution of cancer and the clinical management of patients.94, 95, 96 Although cancer cell populations can be histologically indistinguishable at the microscopical level, they are thought to have unique molecular phenotypes that drive tumor progression and determine the disease outcome of the patient individually.97, 98 The identification of these clinically relevant tumor subpopulations is thus of utmost importance for understanding cancer development and the role of intratumor heterogeneity in the management of cancer patients.99 Because of its ability to perform molecular analyses while retaining morphological information, MALDI Imaging is very well suited to study molecular distribution patterns within a tumor. Some of the first studies focusing on tumor heterogeneity identified protein and lipid intertumoral and intratumoral heterogeneity in myxoid sarcomas.100, 101 In a very recently published proof-of-principle study, spatially resolved, tumor-specific mass spectra were acquired using MALDI Imaging from the tissues of 63 gastric carcinoma and 32 breast carcinoma patients.102 The mass spectra, representing the proteomic heterogeneity within tumor areas, were grouped by a corroborated statistical clustering algorithm in order to obtain segmentation maps of molecularly distinct regions.102 These regions were presumed to represent different phenotypic tumor subpopulations, which was confirmed by linking the presence of these tumor subpopulations to the patients’ clinical data.102 The approach revealed several of the detected tumor subpopulations to be associated with a different overall survival of the gastric cancer patients and the presence of locoregional metastases in patients with breast cancer.102 The published approach demonstrates for the first time de novo identification and characterization of phenotypic tumor subpopulations by MALDI Imaging without any molecular knowledge about the tumors in advance. By defining complex molecular patterns of the tumor subpopulations, it was possible to identify subpopulations correlated to their survival prognosis.102 This work demonstrates the enormous potential of the MALDI Imaging technology applicable to any kind of cancer tissues that exhibit substantial heterogeneity.
POTENTIALS OF MALDI IMAGING MASS SPECTROMETRY IN PATHOLOGY
The application of MALDI mass spectrometry has already been implemented in routinely clinical use by other medical disciplines. The ‘MALDI Biotyper,’ for example, provides the characterization of bacterial cell cultures by collecting molecular profiles and searching a prepopulated database of profile spectra to identify unknown microbes down to species level.103 This technology has received the CE Mark and FDA approval.1 Hundreds of platforms are currently installed in laboratories worldwide.1 The success for this application of MALDI mass spectrometry is due to the performance advantages of the mass spectrometer, and the final solution provides the data to the clinician faster, more cost effectively, and more accurately than the classical microbial approaches.1, 104 This technology demonstrates that the use of multiple molecular markers provides accurate classification of biological samples.1 As the MALDI Biotyper approach is similar to the use of MALDI Imaging in tissue-based experiments, we believe that this concept could also be of great interest for classification of patient samples, eg, for classification of lymphomas or other cancer types or even for the identification of ‘cancer of unknown primary’.
The ongoing technological developments in MALDI Imaging mass spectrometry are very promising. Current developments on mass accuracy now allow the direct identification of masses out of the measurement. The implementation of molecular information into molecular pathway databases or the integration of multidimensional and complex MALDI Imaging data into systems medicine tools makes MALDI Imaging a valuable tool for a deeper understanding of the complexity of tissue and disease. Not only the focus in technological development is put on higher mass accuracy or higher spatial resolution but also the accessibility of new molecular classes is highly promoted. MALDI Imaging itself is already on a high technological level, which is steadily increasing. Numerous publications demonstrate the highly promising advantages for applications by this method. However, there is a great demand for high-quality morphological expertise and medical context foundation in the studies. Molecular histology has been clearly in the field of pathology for a long time. Pathologist have the unique skills to identify tissues that accurately represent the patient's disease and can evaluate molecular information with a great appreciation for cellular context.105 Taking this all together, there is a great demand for input from the expertise of pathologists to the high-quality MALDI Imaging studies. But there must not only be a restriction as co-worker for pathologist in such studies. Rather pathologist are in a unique position having access to high-quality and well-characterized tissue specimen often in conjunction with clinically relevant data. This enables pathologist and pathology itself to generate clinically relevant studies, which could be enriched by extending the traditionally used methodological spectrum in pathology with multi-molecular level in situ data from MALDI Imaging mass spectrometry, bringing tissue-based research to the next level.
References
Norris JL, Caprioli RM . Imaging mass spectrometry: a new tool for pathology in a molecular age. Proteomics Clin Appl 2013;7:733–738.
Norris JL, Caprioli RM . Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev 2013;113:2309–2342.
Caprioli RM, Farmer TB, Gile J . Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 1997;69:4751–4760.
Stoeckli M, Chaurand P, Hallahan DE et al. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med 2001;7:493–496.
Schwamborn K, Caprioli RM . Molecular imaging by mass spectrometry—looking beyond classical histology. Nat Rev Cancer 2010;10:639–646.
Karas M, Kruger R . Ion formation in MALDI: the cluster ionization mechanism. Chem Rev 2003;103:427–440.
Jaskolla TW, Karas M . Compelling evidence for Lucky Survivor and gas phase protonation: the unified MALDI analyte protonation mechanism. J Am Soc Mass Spectrom 2011;22:976–988.
Rompp A, Spengler B . Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol 2013;139:759–783.
Rompp A, Guenther S, Schober Y et al. Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Ed Engl 2010;49:3834–3838.
Rompp A, Guenther S, Takats Z et al. Mass spectrometry imaging with high resolution in mass and space (HR(2) MSI) for reliable investigation of drug compound distributions on the cellular level. Anal Bioanal Chem 2011;401:65–73.
Komatsu M, Murayama Y, Hashimoto H . Protein fragment imaging using ink jet printing digestion technique. Appl Surf Sci 2008;255:1162–1164.
Chaurand P, Latham JC, Lane KB et al. Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: bypassing formalin fixation. J Proteome Res 2008;7:3543–3555.
Cornett DS, Reyzer ML, Chaurand P et al. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods 2007;4:828–833.
Franck J, Arafah K, Barnes A et al. Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: using microspotting. Anal Chem 2009;81:8193–8202.
van Remoortere A, van Zeijl RJ, van den Oever N et al. MALDI imaging and profiling MS of higher mass proteins from tissue. J Am Soc Mass Spectrom 2010;21:1922–1929.
Mainini V, Bovo G, Chinello C et al. Detection of high molecular weight proteins by MALDI imaging mass spectrometry. Mol Biosyst 2013;9:1101–1107.
Emmert-Buck MR, Bonner RF, Smith PD et al. Laser capture microdissection. Science 1996;274:998–1001.
Espina V, Heiby M, Pierobon M et al. Laser capture microdissection technology. Expert Rev Mol Diagn 2007;7:647–657.
Sun N, Ly A, Meding S et al. High-resolution metabolite imaging of light and dark treated retina using MALDI-FTICR mass spectrometry. Proteomics 2014;14:913–923.
Maier SK, Hahne H, Gholami AM et al. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics 2013;12:2901–2910.
McDonnell LA, Walch A, Stoeckli M et al. MSiMass list: a public database of identifications for protein MALDI MS imaging. J Proteome Res 2014;13:1138–1142.
Pote N, Alexandrov T, Le Faouder J et al. Imaging mass spectrometry reveals modified forms of histone H4 as new biomarkers of microvascular invasion in hepatocellular carcinomas. Hepatology 2013;58:983–994.
Djidja MC, Claude E, Snel MF et al. Novel molecular tumour classification using MALDI-mass spectrometry imaging of tissue micro-array. Anal Bioanal Chem 2010;397:587–601.
Groseclose MR, Massion PP, Chaurand P et al. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 2008;8:3715–3724.
Morita Y, Ikegami K, Goto-Inoue N et al. Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray. Cancer Sci 2010;101:267–273.
Quaas A, Bahar AS, von Loga K et al. MALDI imaging on large-scale tissue microarrays identifies molecular features associated with tumour phenotype in oesophageal cancer. Histopathology 2013;63:455–462.
Steurer S, Borkowski C, Odinga S et al. MALDI mass spectrometric imaging based identification of clinically relevant signals in prostate cancer using large-scale tissue microarrays. Int J Cancer 2013;133:920–928.
Steurer S, Seddiqi AS, Singer JM et al. MALDI imaging on tissue microarrays identifies molecular features associated with renal cell cancer phenotype. Anticancer Res 2014;34:2255–2261.
Steurer S, Singer JM, Rink M et al. MALDI imaging-based identification of prognostically relevant signals in bladder cancer using large-scale tissue microarrays. Urol Oncol 2014;32:1225–1233.
Aichler M, Elsner M, Ludyga N et al. Clinical response to chemotherapy in oesophageal adenocarcinoma patients is linked to defects in mitochondria. J Pathol 2013;230:410–419.
Abrass CK . Cellular lipid metabolism and the role of lipids in progressive renal disease. Am J Nephrol 2004;24:46–53.
Lee CH, Olson P, Evans RM . Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 2003;144:2201–2207.
Paradisi G, Ianniello F, Tomei C et al. Longitudinal changes of adiponectin, carbohydrate and lipid metabolism in pregnant women at high risk for gestational diabetes. Gynecol Endocrinol 2010;26:539–545.
Santos CR, Schulze A . Lipid metabolism in cancer. FEBS J 2012;279:2610–2623.
Sparvero LJ, Amoscato AA, Kochanek PM et al. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. J Neurochem 2010;115:1322–1336.
Jackson SN, Wang HY, Woods AS . In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J Am Soc Mass Spectrom 2005;16:2052–2056.
Benabdellah F, Yu H, Brunelle A et al. MALDI reveals membrane lipid profile reversion in MDX mice. Neurobiol Dis 2009;36:252–258.
Thomas A, Charbonneau JL, Fournaise E et al. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal Chem 2012;84:2048–2054.
Rujoi M, Estrada R, Yappert MC . In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal Chem 2004;76:1657–1663.
Rujoi M, Jin J, Borchman D et al. Isolation and lipid characterization of cholesterol-enriched fractions in cortical and nuclear human lens fibers. Invest Ophthalmol Vis Sci 2003;44:1634–1642.
Yappert MC, Rujoi M, Borchman D et al. Glycero- versus sphingo-phospholipids: correlations with human and non-human mammalian lens growth. Exp Eye Res 2003;76:725–734.
Benabdellah F, Seyer A, Quinton L et al. Mass spectrometry imaging of rat brain sections: nanomolar sensitivity with MALDI versus nanometer resolution by TOF-SIMS. Anal Bioanal Chem 2010;396:151–162.
Touboul D, Halgand F, Brunelle A et al. Tissue molecular ion imaging by gold cluster ion bombardment. Anal Chem 2004;76:1550–1559.
Astigarraga E, Barreda-Gomez G, Lombardero L et al. Profiling and imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal Chem 2008;80:9105–9114.
Burnum KE, Cornett DS, Puolitaival SM et al. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res 2009;50:2290–2298.
Colsch B, Woods AS . Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 2010;20:661–667.
Garrett TJ, Dawson WW . Lipid geographical analysis of the primate macula by imaging mass spectrometry. Methods Mol Biol 2009;579:247–260.
Hayasaka T, Goto-Inoue N, Zaima N et al. Organ-specific distributions of lysophosphatidylcholine and triacylglycerol in mouse embryo. Lipids 2009;44:837–848.
Landgraf RR, Prieto Conaway MC, Garrett TJ et al. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal Chem 2009;81:8488–8495.
Meriaux C, Franck J, Wisztorski M et al. Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J Proteomics 2010;73:1204–1218.
Mikawa S, Suzuki M, Fujimoto C et al. Imaging of phosphatidylcholines in the adult rat brain using MALDI-TOF MS. Neurosci Lett 2009;451:45–49.
Murphy RC, Hankin JA, Barkley RM . Imaging of lipid species by MALDI mass spectrometry. J Lipid Res 2009;50 (Suppl):S317–S322.
Snel MF, Fuller M . High-spatial resolution matrix-assisted laser desorption ionization imaging analysis of glucosylceramide in spleen sections from a mouse model of Gaucher disease. Anal Chem 2010;82:3664–3670.
Sparvero LJ, Amoscato AA, Dixon CE et al. Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities and expectations. Chem Phys Lipids 2012;165:545–562.
Sugiura Y, Konishi Y, Zaima N et al. Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res 2009;50:1776–1788.
Vidova V, Pol J, Volny M et al. Visualizing spatial lipid distribution in porcine lens by MALDI imaging high-resolution mass spectrometry. J Lipid Res 2010;51:2295–2302.
Wang X, Han J, Pan J et al. Comprehensive imaging of porcine adrenal gland lipids by MALDI-FTMS using quercetin as a matrix. Anal Chem 2014;86:638–646.
Kurabe N, Hayasaka T, Ogawa M et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci 2013;104:1295–1302.
Miura D, Fujimura Y, Yamato M et al. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal Chem 2010;82:9789–9796.
Hattori K, Kajimura M, Hishiki T et al. Paradoxical ATP elevation in ischemic penumbra revealed by quantitative imaging mass spectrometry. Antioxid Redox Signal 2010;13:1157–1167.
Bao Y, Mukai K, Hishiki T et al. Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol Cancer Res 2013;11:973–985.
Nilsson A, Fehniger TE, Gustavsson L et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One 2010;5:e11411.
Morosi L, Spinelli P, Zucchetti M et al. Determination of paclitaxel distribution in solid tumors by nano-particle assisted laser desorption ionization mass spectrometry imaging. PLoS One 2013;8:e72532.
Pirman DA, Reich RF, Kiss A et al. Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal Chem 2013;85:1081–1089.
Prideaux B, Stoeckli M . Mass spectrometry imaging for drug distribution studies. J Proteomics 2012;75:4999–5013.
Reyzer ML, Caprioli RM . MALDI-MS-based imaging of small molecules and proteins in tissues. Curr Opin Chem Biol 2007;11:29–35.
Reyzer ML, Hsieh Y, Ng K et al. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 2003;38:1081–1092.
Atkinson SJ, Loadman PM, Sutton C et al. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom 2007;21:1271–1276.
Drexler DM, Garrett TJ, Cantone JL et al. Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J Pharmacol Toxicol Methods 2007;55:279–288.
Signor L, Varesio E, Staack RF et al. Analysis of erlotinib and its metabolites in rat tissue sections by MALDI quadrupole time-of-flight mass spectrometry. J Mass Spectrom 2007;42:900–909.
Khatib-Shahidi S, Andersson M, Herman JL et al. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem 2006;78:6448–6456.
Reyzer ML, Chaurand P, Angel PM et al. Direct molecular analysis of whole-body animal tissue sections by MALDI imaging mass spectrometry. Methods Mol Biol 2010;656:285–301.
Rohner TC, Staab D, Stoeckli M . MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev 2005;126:177–185.
Sugiura Y, Setou M . Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: toward in situ pharmacometabolomes. J Neuroimmune Pharmacol 2010;5:31–43.
Prideaux B, Staab D, Stoeckli M . Applications of MALDI-MSI to pharmaceutical research. Methods Mol Biol 2010;656:405–413.
Marko-Varga G, Fehniger TE, Rezeli M et al. Drug localization in different lung cancer phenotypes by MALDI mass spectrometry imaging. J Proteomics 2011;74:982–992.
Sun N, Walch A . Qualitative and quantitative mass spectrometry imaging of drugs and metabolites in tissue at therapeutic levels. Histochem Cell Biol 2013;140:93–104.
Greer T, Sturm R, Li L . Mass spectrometry imaging for drugs and metabolites. J Proteomics 2011;74:2617–2631.
Castellino S, Groseclose MR, Wagner D . MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis 2011;3:2427–2441.
Lietz CB, Gemperline E, Li L . Qualitative and quantitative mass spectrometry imaging of drugs and metabolites. Adv Drug Deliv Rev 2013;65:1074–1085.
Huber K, Feuchtinger A, Borgmann D et al. A novel approach of MALDI drug imaging, immunohistochemistry, and digital image analysis for drug distribution studies in tissues. Anal Chem 2014;86:10568–10575.
Balluff B, Rauser S, Meding S et al. MALDI imaging identifies prognostic seven-protein signature of novel tissue markers in intestinal-type gastric cancer. Am J Pathol 2011;179:2720–2729.
Bauer JA, Chakravarthy AB, Rosenbluth JM et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res 2010;16:681–690.
Schwamborn K, Krieg RC, Jirak P et al. Application of MALDI imaging for the diagnosis of classical Hodgkin lymphoma. J Cancer Res Clin Oncol 2010;136:1651–1655.
Pavlidis N, Fizazi K . Cancer of unknown primary (CUP). Crit Rev Oncol Hematol 2005;54:243–250.
Pavlidis N, Pentheroudakis G . Cancer of unknown primary site. Lancet 2012;379:1428–1435.
Bugat R, Bataillard A, Lesimple T et al. Summary of the Standards, Options and Recommendations for the management of patients with carcinoma of unknown primary site (2002). Br J Cancer 2003;89 (Suppl 1):S59–S66.
Meding S, Nitsche U, Balluff B et al. Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J Proteome Res 2012;11:1996–2003.
Balluff B, Elsner M, Kowarsch A et al. Classification of HER2/neu status in gastric cancer using a breast-cancer derived proteome classifier. J Proteome Res 2010;9:6317–6322.
Rauser S, Marquardt C, Balluff B et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J Proteome Res 2010;9:1854–1863.
Casadonte R, Kriegsmann M, Zweynert F et al. Imaging mass spectrometry to discriminate breast from pancreatic cancer metastasis in formalin-fixed paraffin-embedded tissues. Proteomics 2014;14:956–964.
Lazova R, Seeley EH, Keenan M et al. Imaging mass spectrometry—a new and promising method to differentiate Spitz nevi from Spitzoid malignant melanomas. Am J Dermatopathol 2012;34:82–90.
M'Koma AE, Seeley EH, Washington MK et al. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis 2011;17:875–883.
Dalerba P, Kalisky T, Sahoo D et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011;29:1120–1127.
Gerlinger M, Rowan AJ, Horswell S et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–892.
Maley CC, Galipeau PC, Finley JC et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 2006;38:468–473.
Wu JM, Halushka MK, Argani P . Intratumoral heterogeneity of HER-2 gene amplification and protein overexpression in breast cancer. Hum Pathol 2010;41:914–917.
Greaves M, Maley CC . Clonal evolution in cancer. Nature 2012;481:306–313.
Turner NC, Reis-Filho JS . Genetic heterogeneity and cancer drug resistance. Lancet Oncol 2012;13:e178–e185.
Willems SM, van Remoortere A, van Zeijl R et al. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J Pathol 2010;222:400–409.
Jones EA, van Remoortere A, van Zeijl RJ et al. Multiple statistical analysis techniques corroborate intratumor heterogeneity in imaging mass spectrometry datasets of myxofibrosarcoma. PLoS One 2011;6:e24913.
Balluff B, Frese CK, Maier SK et al. De novo discovery of phenotypic intra-tumor heterogeneity using imaging mass spectrometry. J Pathol 2014;235:3–13.
Bessede E, Angla-Gre M, Delagarde Y et al. Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a University hospital. Clin Microbiol Infect 2011;17:533–538.
Dhiman N, Hall L, Wohlfiel SL et al. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 2011;49:1614–1616.
Berman DM, Bosenberg MW, Orwant RL et al. Investigative pathology: leading the post-genomic revolution. Lab Invest 2012;92:4–8.
Buck A, Halbritter S, Spath C et al. Distribution and quantification of irinotecan and its active metabolite SN-38 in colon cancer murine model systems using MALDI MSI. Anal Bioanal Chem 2015, e-pub ahead of print.
Acknowledgements
This work was supported by the Ministry of Education and Research of the Federal Republic of Germany (BMBF) (Grant No. 01ZX1310B) and the Deutsche Forschungsgemeinschaft (Grant Nos. HO 1258/3-1, SFB 824 TP Z02 and WA 1656/3-1) to AW.
Author information
Authors and Affiliations
Corresponding author
Additional information
Imaging mass spectrometry (IMS) is a novel tissue based research tool that maps the distribution of multiple analytes directly with histological features. Several applications of IMS are discussed such as tissue based disease classification, tumoral heterogeneity, therapy response and prognosis.
Rights and permissions
About this article
Cite this article
Aichler, M., Walch, A. MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab Invest 95, 422–431 (2015). https://doi.org/10.1038/labinvest.2014.156
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2014.156
This article is cited by
-
Metabolic heterogeneity affects trastuzumab response and survival in HER2-positive advanced gastric cancer
British Journal of Cancer (2024)
-
Imaging the metabolic reprograming of fatty acid synthesis pathway enables new diagnostic and therapeutic opportunity for breast cancer
Cancer Cell International (2023)
-
Label-free, multi-parametric assessments of cell metabolism and matrix remodeling within human and early-stage murine osteoarthritic articular cartilage
Communications Biology (2023)
-
Desorption electrospray ionization and matrix-assisted laser desorption/ionization as imaging approaches for biological samples analysis
Analytical and Bioanalytical Chemistry (2023)
-
Moving translational mass spectrometry imaging towards transparent and reproducible data analyses: a case study of an urothelial cancer cohort analyzed in the Galaxy framework
Clinical Proteomics (2022)