Abstract
Stathmin 1 (STMN1), a major microtubule-depolymerizing protein, is involved in cell cycle progression and cell motility. However, the clinical significance of STMN1 expression in non-small cell lung cancer (NSCLC) has not been determined. The expression pattern of STMN1 mRNA was analyzed by quantitative real-time PCR (qRT-PCR) in 37 cases of NSCLC and in the corresponding non-tumor tissue samples. Furthermore, immunohistochemistry was performed to detect STMN1 protein expression in 113 primary NSCLC tissues. The functional role of STMN1 in lung cancer cell lines was evaluated by small interfering RNA-mediated depletion followed by analyses of cell proliferation and invasion. We found that the STMN1 mRNA and protein levels in NSCLC tissues were significantly higher than those in the corresponding non-tumor tissues (P<0.001). In addition, increased STMN1 expression was correlated with poor tumor differentiation (P<0.001), large tumor size (P=0.022), advanced N stage (P=0.033), and advanced TNM stage (P<0.001). Kaplan–Meier analysis indicates that NSCLC patients with higher STMN1 expression showed significantly worse survival. Moreover, multivariate analysis indicates that higher STMN1 protein expression was an independent prognostic factor of disease-specific survival (HR 2.247, 95%CI 1.320-3.825, P=0.003). Finally, the knockdown of STMN1 in lung cancer cells resulted in a decrease in cellular proliferation and invasion. Our findings suggest that STMN1 may have an important role in NSCLC progression and could serve as a potential prognostic marker for patients with NSCLC.
Similar content being viewed by others
Main
Lung cancer is the most common cancer as well as the leading cause of cancer-related deaths around the world.1 The majority of diagnosed lung cancer cases are non-small cell lung cancers (NSCLCs). Despite of the emergence of new cytotoxic drugs and targeted biological agents, NSCLC remains one of the most clinically challenging cancers. Thus, to provide better treatment strategies, there is an urgent requirement to identify new prognostic biomarkers and therapeutic targets for NSCLC.
Stathmin 1 (STMN1) is a major cytosolic phosphoprotein that has a crucial role in the control of cellular division and proliferation by regulating the dynamics of the microtubules.2 In addition to its role in mitosis and cell cycle progression, STMN1 is involved in a wide variety of human cancers, including gastric cancer, nasopharyngeal carcinoma, colorectal cancer, and endometrial carcinoma, etc.3, 4, 5, 6 For example, Kang et al7 showed that STMN1 was upregulated in gastric cancer cell lines and primary gastric adenocarcinomas. Ke et al8 suggest that overexpression of STMN1 was associated with poor prognosis of patients with gastric cancer. Additionally, high STMN1 expression was associated with poor recurrence-free and overall survival in endometrial carcinoma.6 Taken together, these facts indicate that there may be a strong correlation between STMN1 expression and cancer aggressiveness. Chen et al9 suggest that overexpression of STMN1 correlated with poor differentiation in lung adenocarcinomas. However, whether the aberrant expression of STMN1 in NSCLC is associated with malignancy, metastasis, or prognosis remains unknown.
In this study, we examined STMN1 mRNA and protein expressions in NSCLC tissues and adjacent normal tissues by real-time quantitative reverse transcription PCR (qRT-PCR), immunohistochemistry (IHC), and western blotting analysis, respectively. In addition, we assessed the relationship between STMN1 expression and clinicopathological characteristics, and we evaluated its relation with disease-specific survival (DSS) of patients. Finally, we assessed whether STMN1 influenced cell proliferation and invasion in vitro.
MATERIALS AND METHODS
Patients and Specimens
Between 2005 and 2012, 156 NSCLC patients undergoing resection were enrolled in this study from Shanghai Changzheng Hospital. The patients were followed up by phone calls or clinic visits every 6 months until disease progression, death, or study end, except for those who were lost to follow-up (n=8). Follow-up studies included physical examination, laboratory analysis, and computed tomography scan if necessary. Patients who died from diseases other than NSCLC or from unexpected events were excluded from this study (n=35). Finally, a total of 113 NSCLC patients for IHC analysis were enrolled in this study. These NSCLC cases included 72 men and 41 women, with a median age of 55 years. Follow-up lasted to 30 June 2013, with a median follow-up period of 43.4 months for living patients (range: 8.4–88.2 months). During the follow-up time, 75 deaths from NSCLC were observed. In addition, another 37 NSCLC fresh tissues and paired adjacent normal lung tissues from NSCLC patients who had underwent curative resection were collected in 2013 (no follow-up data). None of the patients received preoperative chemotherapy or radiotherapy. Detailed information about demography, clinical characteristics, and histopathology were collected retrospectively for all patients. The histological diagnosis and grade of tumor differentiation were defined through the evaluation of hematoxylin and eosin-stained tissue sections, according to the classification guidelines of the World Health Organization.10 All patients were staged based on the International Association for the Study of Lung Cancer Tumor–Node–Metastasis (TNM) classification, Seventh edition.11 Patients who were included before 2008 were re-reviewed and staged accordingly. Informed consent was obtained from all patients prior to the surgical operations. This study was reviewed and approved by the Ethics Committee of Shanghai Changzheng Hospital. Surgically excised tumors and matched non-cancerous tissues used for qRT-PCR were immediately immersed in liquid nitrogen and stored at −80 °C.
Cell Culture
The lung cancer-derived cell lines used in this study were A549 and H1299 cells. The cells were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum (Invitrogen), 100 IU/ml penicillin (Sigma, St Louis, MO, USA), and 100 mg/ml streptomycin (Sigma). Cells were grown on sterile tissue culture dishes and passaged every 2–3 days using 0.25% trypsin (Invitrogen).
Extraction of Total RNA and Real-Time Quantitative PCR
Total RNA was isolated using the RNA simple total-RNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Reverse transcription was performed in a 25-ml reaction volume with 2 mg total RNA treated with 0.5 mg of Oligo(dt), 200 U M-MLV reverse transcriptase, 25 U RNase inhibitor and 2.5 mM dNTP to synthesize first-strand cDNA (Promega, USA), according to the manufacturer’s instructions. Quantitative RT-PCR was carried out using SYBR Green (Tiangen Biotech) on an Exicycler 96 real-time quantitative thermal block (Bioneer Corporation, Daejeon, Republic of South Korea). The PCR primer sequences were designed according to the human STMN1 and β-actin gene sequences reported in GenBank and were chemically synthesized as follows: for stathmin1, 5′-CCCCTTTCCCCTCCAAAGAA-3′ (forward), 5′-TCGCAAACGTTCCAGTTTGG-3′ (reverse); and for β-actin, 5′-ATCATGTTTGAGACCTTCAA-3′ (forward), 5′-CATCTCTTGCT CGAAGTCCA-3′ (reverse). A melting curve was generated at the end of every run to ensure product uniformity. β-Actin served as the internal control. PCR reactions of each sample were conducted in triplicate. The relative levels of gene expression were represented as ΔCt=Ct gene−Ct reference, and the fold change of gene expression was calculated by the 2−ΔΔCt method. The relative mRNA levels of STMN1 in cancer tissues higher than the mean value (0.870) of relative mRNA levels in adjacent normal lung tissues was defined as high expression.
Immunohistochemistry Analysis
Formalin-fixed and paraffin-embedded specimens were cut into 5-μm thick sections and mounted on glass slides. After dewaxing in xylene and rehydrating stepwise in ethanol, the sections were subjected to heat-induced antigen retrieval. Subsequently, endogenous peroxidase activity and non-specific protein binding were blocked with 3% hydrogen peroxide and 10% normal goat serum, respectively. The sections were then incubated at 4 °C overnight with a rabbit anti-human STMN1 polyclonal antibody (dilution 1:50; Cell Signaling, USA) or anti-p53 antibody (dilution 1:100; Zhongshan). After thoroughly washing with 0.01 mol/l phosphate-buffered saline solution, the corresponding secondary antibody was applied and incubated at room temperature for 30 min. Immunolabeled sections were visualized by using 3,3′-diaminobenzidine, then counterstained with hematoxylin, dehydrated, and mounted. Sections were stained in parallel without primary antibody to provide a negative control.
The immunostaining sections were viewed by two pathologists independently using an Olympus CX31 microscope (Olympus, Japan) in a blind manner. Five views were examined per slide, and 100 cells were observed per view at × 200 magnification. Cells were positive for STMN1 protein when the cell cytoplasm was stained. The signal was quantified by the Allred score system, which represents the estimated intensity and proportion of positive-staining cells.12 The coloring intensity of cells was graded as: ‘1’ (faint yellow), ‘2’ (yellow or deep yellow), and ’3’ (tan or brown). Count was according to the percentage of positive cells: 0=5–25%, 1=26–50%, 2=51–75%, and 3=≥75%. Product of staining intensity and the percentage of positive cells >2 was defined as immune-positive; staining results were divided into four grades according to the product score: ‘−’ (negative, 0∼1), ‘+’ (weakly positive, 2∼3), ‘++’ (moderate positive, 4∼6), and ‘+++’ (strong positive, >6). A score >3 was designated as high expression and a score ≤3 was regarded as low expression.
Western Blotting Analysis
Protein lysates were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). Subsequently, the membrane was incubated with rabbit monoclonal antibody against human STMN1 (1: 500, Cell Signaling Technology) followed by horseradish peroxidase-labeled goat anti-rabbit IgG (1: 2000, Cell Signaling Technology) and detected by chemiluminescence. GAPDH was used as a protein loading control. The intensity of protein fragments was quantified with the ImageJ software.
Small Interfering RNA Treatment
The siRNAs to STMN1 (5′-GAAAGACGCAAGUCCCAUG-3′ and 5′-UAAAGAGAACCGAGAGGCA-3′) and scrambled siRNA were purchased from Genepharma (Shanghai, China). For transfections, cells were seeded in a six-well plate 24 h before the experiment. The cells were transfected with siRNAs using DharmaFECT 1 (0.20 ml/well; ThermoFisher Scientific) according to the manufacturer’s protocol. After 48 h, total RNA and protein and STMN1 expression was measured by qRT-PCR and western blotting as described above.
Cell Proliferation Assay
The cell proliferation assay was performed using the Cell Counting Kit-8 solution (Dojindo, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Briefly, cells were seeded at a concentration of 5 × 103 cells/100 ml/well in 96-well culture plates and treated with 10 ml/well of the Cell Counting Kit-8 solution during the last 4 h of culture. The optical density of the well was measured at 450 nm using a microplate reader.
Matrigel Invasion Assay
One day after transfection with scrambled siRNA or STMN1 siRNAs, cells were trypsinized and then suspended at a density of 5 × 104 cells per ml in 500 ml of RPMI1640 supplemented with 0.5% fetal bovine serum and mitomycin C (0.01 mg/ml, Sigma), added to 8-mm porous BioCoat Matrigel chamber inserts (BD Biosciences, San Jose, CA, USA), and placed in wells filled with 0.7 ml of medium supplemented with 10% fetal calf serum as chemoattractant. After 2 days of incubation, the upper side of the filter was scraped with a cotton tip to eliminate cells that had not migrated through it. The invasive ability of the cells was determined by counting the cells that had migrated to the lower side of the filter with a microscope. Experiments were performed in triplicate, and at least 10 fields were counted in each experiment.
Statistical Analysis
Significant between-group differences were estimated using chi-squared tests, Student’s t-tests, or Fisher’s exact test, as appropriate. DSS was defined as the length of time between the surgery and death specifically from the NSCLC. Kaplan–Meier analysis with the log-rank test was employed to evaluate the relationship between STMN1 expression and DSS. The univariate and multivariate Cox proportional hazards model was utilized to estimate the hazard ratios (HRs) and 95% confidence intervals (CIs) for the outcome. Variables with a value of P<0.05 in univariate analysis (age, gender, smoking status, p53 expression, histology, differentiation, tumor size, N stage, TNM stage, and STMN1 expression) were used in subsequent multivariate analysis on the basis of Cox proportional hazards model. All P-values quoted were two sided, and P<0.05 was considered statistically significant. Statistical analysis was performed using the SPSS v. 18.0 (SPSS, Inc, Chicago, IL, USA).
RESULTS
Overexpression of STMN1 in NSCLC Tissues
First, we quantitatively examined the expression of STMN1 mRNA levels in 37 pairs of primary NSCLC tissues and their corresponding non-tumor samples by using quantitative real-time PCR. Our results show that the relative expression of STMN1 mRNA in NSCLC tissues was significantly higher than in the corresponding adjacent non-tumor tissues (Figure 1a, P<0.001), and 34 out of the 37 patients showed a higher expression level of STMN1 mRNA in NSCLC specimens than in non-cancerous tissue specimens (Figure 1b).
High levels of STMN1 are correlated with progression and prognostic significance in NSCLC. (a) STMN1 expression in cancer tissues and adjacent normal tissues and from NSCLC patients. STMN1 expression was significantly higher in tumor tissue compared with adjacent normal tissue (P<0.001). (b) High STMN1 expression levels were observed in NSCLC tissues in 34 (91.9%) cases. (c) A representative western blotting shows the relative STMN1 protein expression level in tumorous tissue samples of two NSCLC patients (d) The Kaplan–Meier disease-specific survival curves by STMN1 levels. Patients with elevated STMN1 expression showed reduced survival times compared with patients with low levels of STMN1 expression (log-rank test; P<0.001).
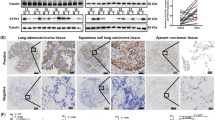
IHC was employed to examine the protein expression of STMN1 in primary NSCLC specimens and normal adjacent tissues. Immunoreactivity of STMN1 was observed primarily in the cytoplasm (Figure 2). The samples were classified into four groups: group 1 with negative staining (−), group 2 with weak staining (+), group 3 with the moderate staining (++), and group 4 with strong staining (+++) (Figures 2a–d). Groups 1 and 2 were regarded as low STMN1 protein expression (score ≤3), while groups 3 and 4 were regarded as high STMN1 protein expression (score >3). After exclusion of cases that could not be evaluated due to tissue loss or inadequate tissue, the positive rates of STMN1 expression in normal tissues was 12.3%, and the positive rates of STMN1 in NSCLC was 87.9%. The protein level of STMN1 in NSCLC was significantly higher than that expressed in normal adjacent tissues (P<0.001).
Negative and positive staining of STMN1 protein expression in human NSCLC and normal adjacent tissues by IHC. Negative and positive staining of STMN1 protein in normal and tumor tissues were classified into four groups (− to +++) according to the staining intensity and extension of each tissue. Negative staining (−) and weak staining (+) were regarded as low STMN1 protein expression (score ≤3), while moderate staining (++) and strong staining (+++) were regarded as high STMN1 protein expression (score >3).
Correlation of STMN1 Expression with Clinicopathological Parameters
According to the results of IHC, 61 out of the 113 patients were classified as low STMN1 group (score <3) and 52 out of the 113 patients were classified as high STMN1 group (score ≥3). By randomly selecting some fresh cancer tissues and adjacent normal tissues for western blotting analysis of STMN1 protein, we found that upregulated STMN1 protein expression was observed in the high score group comparing with the low score group (Figure 1c). We then evaluated the association between STMN1 protein expression and the clinicopathological data of the patients. As summarized in Table 1, we found that a high expression of STMN1 protein was significantly associated with poor tumor differentiation (P<0.001), large tumor size (P=0.022), advanced N stage (P=0.033), and advanced TNM stage (P<0.001). No significant association was found between STMN1 protein expression and other clinical features, including gender, age, smoking status, p53 expression, and histology.
Association of STMN1 expression with prognosis of NSCLC
The Kaplan–Meier method was used to analyze the association of STMN1 protein expression with prognosis of NSCLC patients. We found that the DSS of patients with high STMN1 protein expression were significantly shorter than that of patients with low STMN1 protein expression (HR 3.205, 95% CI 1.849–5.263, P<0.001; Figure 1d).
To evaluate the possibility of STMN1 as an independent risk factor for poor prognosis, conventional clinicopathological factors and STMN1 protein levels were assessed by Cox’s univariate and multivariate hazard regression model (Table 2). Univariate analysis indicates that tumor differentiation, N stage, TNM stage, and high STMN1 protein expression were significantly associated with DSS of NSCLC patients. Variables with a value of P<0.05 were selected for multivariate analysis. Multivariate analysis showed that high STMN1 protein expression (HR 2.247, 95% CI 1.320–3.825, P=0.003) and TNM stage (HR 4.326, 95% CI 2.343–7.988, P<0.001) were independent prognostic factors for DSS of NSCLC patients.
Role of STMN1 in Lung Cancer Cells
To explore the biological function of STMN1 in lung cancer cells, we employed siRNAs to knockdown STMN1 expression in both A549 and H1299 cell lines. When A549 and H1299 cells were transfected with STMN1 siRNAs, the expression of STMN1 was reduced in both the mRNA and protein levels (Figures 3a and b). STMN1 depletion significantly affected the proliferation of A549 (Figure 3c) or H1299 (Figure 3d) cells. We next examined the role of STMN1 in cancer cell invasion. We observed a significant difference between cells transfected with scrambled siRNA and STMN1 siRNA in the Matrigel invasion assay (Figure 4). Taken together, these results indicate that STMN1 contributed to the progression of NSCLC in vitro.
Silencing STMN1 inhibits the proliferation of NSCLC cells. Silencing STMN1 in two siRNA-transduced stable NSCLC cell lines. Relative gene expression determinations were made using the qRT-PCR (a) and western blotting (b) analysis. The A549 (c) or H1299 (d) cells were transfected with scrambled siRNA or two individual siRNAs against STMN1 as indicated. Cell viability was detected at the indicated time points after transfection using the CCK-8 assays. (*P<0.05).
Silencing STMN1 inhibits the metastasis of NSCLC cells. The representative pictures (top panel) and quantification (bottom panel) of invaded cells were analyzed using the Transwell matrix penetration assay. Significant reduction in the invasive ability was shown on STMN1 knockdown (*P<0.05). The cell number was counted in five random view fields, and the error bars represent s.d.s.
DISCUSSION
In this study, we report that the relative level of STMN1 mRNA expression in primary NSCLC tissues was significantly higher than that in the corresponding non-tumor lung tissues. High level STMN1 protein expression was also observed in 52 out of the 113 NSCLC patients. We then showed that high level of STMN1 protein was significantly associated with tumor differentiation, tumor size, N stage, and TNM stage. Furthermore, statistical analysis suggests that patients with higher STMN1 expression had shorter DSS, whereas patients with lower STMN1 expression had a better survival. We also found an impaired proliferation capacity and invasive ability after STMN1 knockdown.
STMN1 expression and activity have been examined in several types of cancer. Most of the studies were concordant with the notion that STMN1 expression was upregulated in human cancers. For example, the expression of STMN1 was elevated in NSCLC.9 Our data also confirmed this finding. More importantly, it was to note that STMN1 overexpression has been associated with poor survival of cancers, including colorectal cancer, hepatocellular carcinoma, and oral squamous-cell carcinoma.5, 9, 13, 14 We demonstrated that overexpression of STMN1 was an independent prognostic factor for survival in patients with NSCLC.
A previous study reported that there was an increased STMN1 expression in the resistant A549 cell line,15 and the expression level of STMN1 was negatively correlated with the efficacy of vinorelbine plus cisplatin/carboplatin therapy in late NSCLC.16 In addition, Rosell et al17 found that STMN1 levels could influence the time to progression of vinorelbine/cisplatin-treated patients. Taken together, these results indicate that STMN1 may be correlated with tumor drug resistance and chemosensitivity. However, Jiang et al18 found that there was no significant association between the mRNA expression of STMN1 and chemosensitivity of patients with NSCLC. Whether the differences were due to different population from different region and race or due to the different procedures used to evaluate the STMN1 expression (qRT-PCR vs IHC) was not clear. Thus more studies are needed to investigate this issue in the future.
STMN1 expression in human cancer was almost invariably associated with increased local invasion and metastasis formation, independently of the histology of the primary tumor.19, 20 In this study, overexpression of STMN1 was found to closely link to tumor differentiation, tumor size, N stage, and TNM stage. This result suggests that STMN1 could have a major role in tumor progression and dissemination. Therefore we examined the effect of STMN1 knockdown in A549 and H1299 cell lines. We showed that STMN1 regulated proliferation of lung cancer cells. This may explain why high STMN1 expression was significantly associated with tumor size. In addition, decreasing STMN1 expression significantly enhanced cell migration and invasion, which may explain the greater lymph node metastasis and poor prognosis in NSCLC patients with STMN1 overexpression.
Kang et al7 found that aberrant p53 immunoreactivity was associated with higher STMN1 expression in gastric adenocarcinoma, suggesting that overexpression of STMN1 might be, in part, due to inactivation of tumor-suppressor gene p53. However, no significant association between p53 expression and STMN1 was observed in our study, indicating that there might be other mechanisms involved in the dysregulation of STMN1 in NSCLC. This important issue is needed to be analyzed by functional experiments in vivo and in vitro. In addition, further studies should be designed to determine whether overexpression of STMN1 is an early or late event in lung tumorigenesis and what the underlying mechanism of overexpression is. Results from this study showed that STMN1 was upregulated in the tumor-promotion pathway and that it acted to increase cell growth and invasion. Pharmacological suppression of STMN1 may represent a promising approach for NSCLC treatment.
This study had some limitations. First, although IHC analysis could be used for semi-quantitative evaluation, western blotting carried out on fresh samples may be better for accurate protein quantitative analysis. Second, mRNA and protein analysis were not carried out on the samples from healthy subjects. The frequency of positivity would be useful to know and should be detected henceforth.
In conclusion, we determined that STMN1 expression was significantly overexpressed in NSCLC. Upregulated expression of STMN1 was correlated with poor tumor differentiation, large tumor size, advanced N stage, and advanced TNM stage. STMN1 may have a key role in NSCLC proliferation and metastasis and could be a predictor for DSS in NSCLC patients. Our data suggest that STMN1 might represent a novel prognostic indicator in NSCLC and a potential target for gene therapy.
References
Siegel R, Naishadham D, Jemal A . Cancer statistics, 2013. CA Cancer J Clin 2013;63:11–30.
Belmont LD, Mitchison TJ . Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 1996;84:623–631.
Jeon TY, Han ME, Lee YW et al. Overexpression of stathmin1 in the diffuse type of gastric cancer and its roles in proliferation and migration of gastric cancer cells. Br J Cancer 2010;102:710–718.
Hsu HP, Li CF, Lee SW et al. Overexpression of stathmin 1 confers an independent prognostic indicator in nasopharyngeal carcinoma. Tumour Biol 2014;35:2619–2629.
Zheng P, Liu YX, Chen L et al. Stathmin, a new target of PRL-3 identified by proteomic methods, plays a key role in progression and metastasis of colorectal cancer. J Proteome Res 2010;9:4897–4905.
Salvesen HB, Carter SL, Mannelqvist M et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci USA 2009;106:4834–4839.
Kang W, Tong JH, Chan AW et al. Stathmin1 plays oncogenic role and is a target of microRNA-223 in gastric cancer. PLoS One 2012;7:e33919.
Ke B, Wu LL, Liu N et al. Overexpression of stathmin 1 is associated with poor prognosis of patients with gastric cancer. Tumour Biol 2013;34:3137–3145.
Chen G, Wang H, Gharib TG et al. Overexpression of oncoprotein 18 correlates with poor differentiation in lung adenocarcinomas. Mol Cell Proteomics 2003;2:107–116.
Brambilla E, Travis WD, Colby TV et al. The new World Health Organization classification of lung tumours. Eur Respir J 2001;18:1059–1068.
Goldstraw P, Crowley J, Chansky K et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706–714.
Allred DC, Clark GM, Elledge R et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst 1993;85:200–206.
Hsieh SY, Huang SF, Yu MC et al. Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Mol Carcinog 2010;49:476–487.
Yuan RH, Jeng YM, Chen HL et al. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol 2006;209:549–558.
Martello LA, Verdier-Pinard P, Shen HJ et al. Elevated levels of microtubule destabilizing factors in a Taxol-resistant/dependent A549 cell line with an alpha-tubulin mutation. Cancer Res 2003;63:1207–1213.
Pu X, Wang J, Xu L et al. Relationship between expression of beta-tubulin-III plus stathmin in advanced non-small cell lung cancer and its sensitivity to venorelbine chemotherapy. Zhongguo Fei Ai Za Zhi 2009;12:49–53.
Rosell R, Scagliotti G, Danenberg KD et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene 2003;22:3548–3553.
Jiang H, Yu XM, Zhou XM et al. Correlation between microtubule-associated gene expression and chemosensitivity of patients with stage II non-small cell lung cancer. Exp Ther Med 2013;5:1506–1510.
Kouzu Y, Uzawa K, Koike H et al. Overexpression of stathmin in oral squamous-cell carcinoma: correlation with tumour progression and poor prognosis. Br J Cancer 2006;94:717–723.
Belletti B, Baldassarre G . Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets 2011;15:1249–1266.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Stathmin 1 (STMN1) is a microtubule-depolymerizing protein involved in cell cycle progression and motility. This paper shows that STMN1 is overexpressed in non-small cell lung cancer (NSCLC). High STMN1 expression correlates with poor tumor differentiation, large tumor size and advanced stages. Knockdown of STMN1 in lung cancer cells decreases proliferation and invasion. Therefore, STMN1 may be prognostic marker for NSCLC.
Rights and permissions
About this article
Cite this article
Nie, W., Xu, Md., Gan, L. et al. Overexpression of stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Lab Invest 95, 56–64 (2015). https://doi.org/10.1038/labinvest.2014.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2014.124
This article is cited by
-
Stathmin 1 is a biomarker for diagnosis of microvascular invasion to predict prognosis of early hepatocellular carcinoma
Cell Death & Disease (2022)
-
Post-translational modifications of CDK5 and their biological roles in cancer
Molecular Biomedicine (2021)
-
Zyxin (ZYX) promotes invasion and acts as a biomarker for aggressive phenotypes of human glioblastoma multiforme
Laboratory Investigation (2020)
-
Phosphorylation of adducin-1 by cyclin-dependent kinase 5 is important for epidermal growth factor-induced cell migration
Scientific Reports (2019)
-
The effect of downregulation of Stathmin gene on biological behaviors of U373 and U87-MG glioblastoma cells
Biological Research (2018)