Abstract
The KIAA0101 protein is overexpressed in various human cancers, including esophageal cancer (EC). This study assessed the association of KIAA0101 protein with prognosis and resistance to chemotherapy in EC patients and then explored the role of KIAA0101 in EC cells in vitro. A total of 228 EC patients participated in the study. Tissue samples were collected for immunohistochemical analysis of KIAA0101 expression in tumor and normal tissues for association with clinicopathological and survival data. KIAA0101 cDNA or shRNA were transfected into EC cells for assessment of tumor cell viability, sensitivity to cisplatin treatment, and gene expression. Array-based comparative genomic hybridization (aCGH) was used to detect the changed copy-number alterations in cell lines expressing different levels of KIAA0101. Expression of KIAA0101 protein was upregulated in EC tissues, which was associated with pTNM stage, resistance to chemotherapy, tumor recurrence, and poor survival of EC patients. In vitro experiments showed that expression of KIAA0101 enhanced cell proliferation and upregulated cyclins A and B expression, leading to a reduced G1 phase of the cell cycle. KIAA0101 also induced resistance of EC Eca-109 and TE-1 cell lines to cisplatin treatment through a decrease in apoptosis. The aCGH data showed that levels of KIAA0101 expression altered chromosome stability, affecting genes that are associated with cancer progression. In conclusion, upregulated KIAA0101 expression is associated with EC progression, resistance to chemotherapy, and poor survival of the patients.
Similar content being viewed by others
Main
Esophageal cancer (EC) is a significant worldwide health problem. Approximately 90% of ECs are squamous cell carcinomas (esophageal squamous cell carcinoma (ESCC)), although esophageal adenocarcinomas account for 60–70% of EC cases in Western countries.1 These two histology types of EC have relatively different risk factors and prevalence rates in different geographic regions, but all have very poor prognosis mainly due to diagnosis at advanced stages.2 To date, surgical resection is the primary treatment, as most patients do not respond to chemotherapy. Thus there is an urgent need for the identification of novel biomarkers for the early detection of EC and/or prediction of responses to treatment.
KIAA0101 is a 15-kDa proliferation cell nuclear antigen (PCNA)-associated protein3 and also known as p15PAF, L5, and OEATC-1. KIAA0101 is involved in the regulation of DNA repair and cell proliferation, cell cycle progression, and migration.4 Overexpression of KIAA0101 protein is able to protect cells from UV-induced cell death.5 During development, KIAA0101 protein expression is highly restricted in a spatiotemporal manner in mouse embryos, whereas it shows aberrant expression in various cancers, such as breast, uterine cervix, brain, kidney, hepatic, lung, esophageal, and colon cancers.6, 7 However, not all of the clinical studies on KIAA0101 have supported the conclusion that KIAA0101 functions as an oncogene in tumor progression,6, 8 perhaps because some functions of the KIAA0101 protein are related to DNA repair; alteration of the latter activity is usually associated with tumorigenesis, but gaining functions of DNA repair will promote tumor progression and resistance to cisplatin-based chemotherapy.9 During cancer development, the lack of KIAA0101 activity alters chromosome instability, for example, changes in gene copy number are noted.9 In contrast, after tumors have developed, expression of KIAA0101 may contribute to chemoresistance in human cancers.
The purpose of the current study is to elucidate the clinicopathological significance of elevated KIAA0101 expression in EC, in particular, its association with prognosis of EC patients and resistance to chemotherapy. We present in vitro studies that investigate the effects of KIAA0101 expression, or knockdown, on the regulation of tumor cell viability, resistance to chemotherapy, and changes in gene copy numbers in EC cells.
MATERIALS AND METHODS
Tissue Samples
A total of 228 surgically resected, unifocal, primary human EC tumor samples were prospectively collected from The Oncology Surgery Department of The First Affiliated Hospital of Xi’an Jiaotong University between January 2005 and January 2009. All 228 tissue samples with matched distant non-cancerous esophageal tissues were histologically diagnosed and snap-frozen in liquid nitrogen after resection and stored in liquid nitrogen until use. Tumor stages and histological grades were recorded using the classification guidelines of the American Joint Committee on Cancer (AJCC), Seventh Edition (2010) published by Springer Science and Business Media LLC. Of these 228 patients, 214 patients with EC were surgically treated with the same therapeutic strategy: complete tumor resection with negative margins (R0 resection) and extensive lymphadenectomy, whereas the remaining 14 cases were at stage IV of the disease and underwent palliative surgery. Further, all patients after surgery accepted postoperative chemotherapy with 5-Fu and leucovorin (Leucovorin 20 mg/m2 on days 1–5, 5-Fu 425 mg/m2 daily on days 1–5 cycled every 28 days) and radiotherapy of 45–50 Gy (1.8 Gy/day). Patients with only palliative surgery or recurrent disease were treated with ECF adjunctive treatment (ie, 50 mg/m2 Epirubicin on day 1, 60 mg/m2 cisplatin on day 1, 200 mg/m2 5-FU continuous infusion over 24 h daily on days 1–21 for three cycles). All patients were followed up on a regular basis for >36 months or until death. The last follow-up was conducted in December 2012. This study was approved by the Committee for the Conduct of Human Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University, and all 228 patients signed written informed consent forms.
Immunohistochemistry
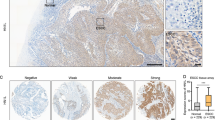
All tissue specimens were fixed in 10% buffered formalin and embedded in paraffin for preparation of consecutive 4-μm-thick sections. For immunohistochemistry, the tissue sections were first de-paraffinized and rehydrated and then subjected to an antigen retrieval treatment with a pressure cooker and incubated with normal serum to block potential nonspecific binding of the secondary antibody. After that, the sections were incubated with a primary mouse anti-human anti-KIAA0101 antibody (Cat. No. H0009768-M01 from Abnova, Taipei, Taiwan) at a dilution of 1:500 at 4 °C overnight. On the following day, the sections were washed with phosphate-buffered saline (PBS) and further incubated with a secondary goat anti-mouse IgG at a dilution of 1:200 (Pierce Biotechnology, Rockford, IL, USA) for 30 min at room temperature and then with 3,3-diaminobenzidine as a chromogen. The sections were finally counterstained with hematoxylin solution and then dehydrated and covered with coverslips. All stained sections were independently evaluated by two investigators, and agreement was reached after careful discussion, if discrepancies occurred. KIAA0101 was scored by adding the percentage of positive tumor cells and staining intensity as described in our previous studies,10 ie, the percentage of positive cells was scored as: 0, <15%; 1+, 16–30%; 2+, 31–60%; and 3+, 61–100%. Intensity of staining was graded as follows: grade 0, negative; 1+, weak positive; 2+, moderate positive; and 3+, strong positive. According to these immunohistochemical scores, KIAA0101 expression was then divided into two groups: negative or weak KIAA0101 expression (score≤3) and positive or strong KIAA0101 expression (score≥4) as shown in Figure 1. Images of the stained sections were obtained using a light microscope (BX51; Olympus, Tokyo, Japan) equipped with a digital camera (PD71; Olympus).
Immunohistochemical analysis of KIAA0101 expression in tissue samples. Positive staining is indicated by a brown nuclear color. (a, c, e, g). Percentage of positive cells was scored as: 0, <15% (a); 1+, 16–30% (c); 2+, 31–60% (e); and 3+, 61–100% (g). (b, d, f, h). Intensity of staining was scored as grade 0 (b), negative; 1+ (d), weak positive; 2+ (f), moderate positive; and 3+, strong positive (h). The sum score was divided into negative or weak KIAA0101 expression at score ≤3 (eg, a, b, c, and d) and positive or strong KIAA0101 expression at score≥4 (eg, e, f, g, and h). (i) Immunohistochemical analysis of one of the normal esophageal tissues.
Cell lines, Culture, and Gene Transfection
Human EC Eca-109 and TE-1 cell lines were obtained from The Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and grown in RPMI-1640 medium (Gibco, New York, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Utah, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco) at 37 °C in a 5% CO2 incubator. To manipulate gene expression in the cells, plasmids carrying KIAA0101 cDNA or shRNA were separately transfected into these two EC cell lines using Xtreme HP (Roche, Mannheim, Germany) according to the manufacturer’s instructions. After 48 h transfection, G418-sulfate (600 ng/μl for Eca-109, and 800 ng/μl for TE-1, Sigma-Aldrich, St Louis, MO, USA) was added to the cell culture medium throughout the experimental period. The cells were fed three times weekly and periodically assessed by western blot to ensure transgene expression.
Construction of Plasmids
To induce expression of the KIAA0101 protein, a full length of KIAA0101 cDNA was cloned into an expression vector p-EGFP. To knock down endogenous KIAA0101 expression in EC cells, we used pSIREN-Shuttle vector to express a short hairpin RNA against KIAA0101 as described in our previous study.10 For negative controls, empty p-EGFP and pSIREN-Shuttle vectors were used in parallel experiments.
Protein Extraction and Western Blot
Cells were lysed with a modified RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) containing 25 μg/ml leupeptin, 1 mM sodium orthovanadate, 2 mM EDTA, and 1 mM PMSF. The concentration of the protein samples was determined using the BCA method (Beyotime, Beijing, China). Twenty microgrm of each protein sample was loaded onto 8% SDS-polyacrylamide gel, electrophoresed, and blotted onto a PVDF membrane (Sigma-Aldrich). The membranes were blotted with the first antibody overnight at 4 °C (ie, Anti-KIAA0101, Anti-cyclins A, B1, C, D and E, and Anti-β-actin antibody, all from Abnova) and with a secondary antibody at room temperature for 1 h. Immunoreactivity was detected using an ECL system (Xian Jiaotong University) and normalized to β-actin.
Cell Viability Assay
Cell viability was assessed by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide MTT assay. Cells with stable-transfected KIAA0101 and 48 h of transfection with shRNA were seeded into 96-well plates at a density of 3000 cells per well and then treated with or without cisplatin at 0.1 mg/l for up to 3 days (Qilu Pharmaceutical, China). At the end of the experiments, 20 μl of MTT (0.5 mg/ml in PBS) were added to the cells, they were then incubated for 4 h at 37 °C, the growth medium was discarded, and 100 μl dimethylsulfoxide was added into each well. After agitation for 10 min on a shaker, absorbance rate was read at 550 nm with a scanning microtiter (PerkinElmer, MA, USA). Data were summarized as percentage of control as compared with the untreated cells.
Cell Cycle and Apoptosis Analyses
The cell cycle distribution of cells was analyzed using propidium iodide staining and flow cytometric analysis. Apoptosis in these cells treated with or without cisplatin was evaluated by flow cytometric techniques using an Annexin-V-FLUOS Staining Kit (Roche) according to the manufacturer’s instructions.
Array Comparative Genomic Hybridization
To detect the effects of KIAA0101 alteration on the regulation of DNA repair capacity as altered chromosome copy numbers, we performed an Array Comparative Genomic Hybridization (aCGH) analysis with the assistance of Kangchen Bio-tech (Shanghai, China). Briefly, 10 million Eca-109-KIAA0101 and Eca-109-shKIAA0101 cells were subjected to genomic DNA extraction using a genomic DNA extraction kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and sonicated to a size of about 200–1000 bp. The shearing DNA was then labeled with Cy3- and Cy5-random 9-mers, respectively, and hybridized to Human CGH 385 K Whole-Genome Tiling Array, which is a single array design containing all chromosomes with 7073 bp median probe spacing and totally covered by ∼385 000 probes. Genomic DNA from parental Eca-109 cells was used as controls to normalize the data. The array was scanned using an Axon GenePix 4000B microarray scanner (Molecular Devices, Union City, MA, USA). Next, the raw data were extracted as pair files by NimbleScan software. Log 2 ratio data were calculated and normalized by spatial correction and fit normalization. After that, the data were analyzed using segments with specified parameters (Min. segment difference of 0.1; Min. segment length (number of probes) of 1; Acceptance percentile of 0.999; Number of permutations of 10; Averaging windows of 10 × and 1 × ), and genomic transcripts were then mapped and genes included by filtered segments where they were shown.
Statistical Analysis
We performed the χ2 test to compare expression of KIAA0101 with clinicopathological data from the patients. We then utilized Spearman and logistic regression analyses to compare KIAA0101 expression between tumor and adjacent normal tissues and Kaplan–Meier curve and logical rank test to analyze survival predictor data of the patients. All in vitro data represent the mean of at least three individual experiments±s.e. and statistically analyzed using the Student’s t-test. All statistical analyses were conducted using the SPSS 13.0 software (SPSS Chicago, IL). A P value <0.05 was considered as statistically significant.
RESULTS
Expression of KIAA0101 Protein Associated with Poor Clinical Outcome of EC Patients
In this study, we detected the expression of KIAA0101 protein in a total of 228 ECs and the corresponding normal tissues using immunohistochemical analysis. We found that expression of the KIAA0101 protein was upregulated in EC tissues compared with the paired non-cancerous tissues (Figure 1), ie, KIAA0101 protein was expressed in 106 of the 228 (46.5%) EC tissues, whereas only 28 of the 228 (12.3%) matched adjacent non-cancer gastric tissues had KIAA0101 staining (P<0.001). We then associated KIAA0101 expression with clinicopathological data from EC patients and found that KIAA0101 expression was associated with pTNM stage (P=0.04), chemotherapy resistance (P<0.001), and tumor recurrence (P<0.001). In addition, KIAA0101 expressed more frequently in advanced-staged cancer tissues. KIAA0101 was expressed in 52 of the 130 (40%) stage I and II tumor tissues but was in 54 of the 98 (55.1%) stage III and IV tumor tissues (P=0.024, Table 1). After that, we separated 206 squamous cell carcinomas from 21 adenocarcinomas for further analysis and found that the elevated KIAA0101 was strongly associated with resistance to chemotherapy and early recurrence (Table 2).
Moreover, we performed a Kaplan–Meier curve analysis, and the data showed that KIAA0101 expression was associated with poor overall (P<0.001) and disease-free survival (P=0.004) for EC patients (Figure 2). In addition, survival was affected by tumor invasion (P=0.001), metastasis (P<0.001), and chemotherapy (P=0.002) based on the log-rank survival analysis (Tables 1 and 3; Figure 2). Multivariate stepwise logistic regression analyses showed that the expression of KIAA0101 protein, tumor pTNM stages, and sensitivity to chemotherapy were all independent predictors for survival of EC patients (Table 3). On the other hand, in squamous cell carcinoma KIAA0101 was associated with poor prognosis (P<0.001), tumor invasion (P=0.008), metastasis (P<0.001), and chemotherapy resistance (P=0.007).
Kaplan–Meier curves of overall and disease-free survival of oesophageal cancer patients. (a) Overall survival stratified by overexpression of KIAA0101 in tumor tissues. (b) Disease-free survival stratified by overexpression of KIAA0101 in tumor tissues. (c) Sensitivity of KIAA0101 expression to chemotherapy. The data showed that upregulated KIAA0101 expression predicted the worse response to chemotherapy.
KIAA0101 Expression Promotes EC Cell Viability by Induction of Cyclins A and B Proteins
Next, we assessed the effects of KIAA0101 expression on regulation of EC cells by transfection of a plasmid carrying KIAA0101 cDNA into EC cells. Our data showed that KIAA0101 expression induced tumor cell viability, whereas knockdown of KIAA0101 expression using KIAA0101 shRNA reduced cell viability by 83.2% in Eca-109 and by 89.2% in TE-1 cells (P<0.05; Figure 3). At the molecular level, expression of the KIAA0101 protein dramatically induced cyclins A and B1 expression but reduced their expression after KIAA0101 knockdown (Figure 4), which was further supported by an increase in G1 phase of the cell cycle after KIAA0101 knockdown, while cell cycle G2 and S phase was not affected parallel to KIAA0101 expression (Figure 3c), but the cells in G2 plus S phase were parallel to its expression, indicating that overexpression of KIAA0101 promoted cell viability by induction of cyclins A and B expression. We also analyzed other genes, such as cyclins C, D and E, but there was no change after altered KIAA0101 expression (Figure 4).
Effects of KIAA0101 expression and knockdown on regulation of esophageal cancer cell viability. (a, b) MTT assay. Esophageal cancer Eca-109 and TE-1 cells were seeded and transfected with KIAA0101 cDNA or shRNA, respectively, and then subjected to MTT assay. (c) Flow cytometric cell cycle assay. The cells were analyzed by flow cytometry.
Expression of KIAA0101 Reduced Cancer Cell Sensitivity to Cisplatin Treatment
As KIAA0101 protein is able to regulate DNA repair,11, 12 we determined whether KIAA0101 expression can affect tumor cell response to cisplatin treatment. After expression of KIAA0101 protein in EC cells, treatment of tumor cells with 0.05 mg/ml cisplatin for 72 h showed that there was a 38, 65, and 24% decrease in cell viability in parental, KIAA0101 shRNA, or KIAA0101cDNA-transfected Eca-109 cells, respectively, and a 32, 44, and 27% decrease in parental, KIAA0101 shRNA, and KIAA0101 cDNA-transfected TE-1 cells, respectively (P<0.05; Figure 5a). These results support the conclusion that elevated KIAA0101 expression enhanced cancer cell resistance to cisplatin-induced cell growth inhibition.
Effects of KIAA0101 expression and knockdown on regulation of tumor cell sensitivity to cisplatin treatment. (a, b) MTT assay. Esophageal cancer Eca-109 and TE-1 cells were seeded and transfected with KIAA0101 cDNA or shRNA, respectively, treated with cisplatin, and then subjected to MTT assay. The data showed that overexpression of KIAA0101 alleviated induced cell growth inhibition, while knockdown of KIAA0101 led cells to be more sensitive to cisplatin treatment at 48 and 72 h (P<0.05). (c) Flow cytometric apoptosis assay. The duplicated cells were subjected to flow cytometry.
Furthermore, the tumor cell apoptosis assay showed that the average apoptotic cell fraction was significantly increased upon knock down of KIAA0101 expression compared with parental cells and KIAA0101-expressed cells (Figure 5c). After being treated with cisplatin for 24 h, knockdown of KIAA0101 expression resulted in about a 3.7-fold and 3.5-fold increase in apoptosis in Eca-109 and TE-1 cells, respectively, compared with the parental controls. This finding suggests that KIAA0101 has a role in tumor cell resistance to cisplatin-induced apoptosis.
KIAA0101 Expression Associated with Altered DNA Copy Numbers in EC Cells
We also performed an aCGH analysis to explore the effects of KIAA0101 knockdown and overexpression on the regulation of chromosome copy numbers. We found that EC cell Eca-109 transfected with KIAA0101 shRNA had 2366 genes with altered copy numbers compared with KIAA0101-overexpressed cells, which had 1255 alterations. After setting the cutoff points of two folds for upregulation and 50% for downregulation, we generalized that 28 genes were gained and 18 genes were lost after upregulation of KIAA0101 (Table 4), some of which were associated with tumor progression. Specifically, copy numbers of Bax, caspase-3, and MUC-2 had the most significant changes after KIAA0101 alteration in Eca-109 cells and these genes are related to cell apoptosis and adhesion.
DISCUSSION
In this study, we analyzed expression of KIAA0101 protein for association with clinicopathological data and then performed in vitro experiments to verify the role of KIAA0101 in ESCC. Our data showed that KIAA0101 protein was highly expressed in ESCC and KIAA0101 expression was associated with pTNM stage, chemotherapy resistance, and tumor recurrence. Moreover, KIAA0101 expression was associated with poor overall and disease-free survival of EC. The multivariate logistic regression analysis showed that expression of KIAA0101 protein, tumor pTNM stages, and sensitivity to chemotherapy were all independent predictors for survival of EC patients. After which we performed in vitro studies and found that KIAA0101 expression induced tumor cell viability, whereas knockdown of KIAA0101 expression reduced cell viability. KIAA0101 expression also reduced tumor cell sensitivity to cisplatin treatment. KIAA0101 knockdown altered chromosome copy numbers in ESCC cells. We propose that further studies to evaluate KIAA0101 protein as a biomarker for the prediction of EC progression and prognosis would be worthwhile.
Indeed, accumulating evidence indicates that KIAA0101 shares a PCNA-binding motif with many cell cycle regulatory factors and has a role in cell cycle progression.13 KIAA0101 enables tumor progression in several cancer types.14 Our previous study showed that the KIAA0101 protein was elevated in gastric cancer tissues and promoted tumor cell proliferation, invasion, and served as a biomarker for prediction of early tumor recurrence.10 However, earlier studies of KIAA0101 expression in EC did not reveal such a role of KIAA0101 in vitro or in vivo.4 In the current study, we proved the conclusion of the previous work,4 confirming that KIAA0101 was elevated in esophageal cancer tissue compared with adjacent tissue, and furthermore using the three types of data: (i) Immunohistochemical analysis using an anti-KIAA0101 antibody validated a higher level of KIAA0101 expression in EC tissue than in gastric cancer, but a low or moderate level of KIAA0101 expression in normal tissues. This finding illustrates the role of KIAA0101 in EC development. (ii) High expression of KIAA0101 protein was associated with tumor early recurrence, short survival, and resistance to cisplatin-based chemotherapy in vitro and in vivo, which was similar to the data on some other cancer types. This finding indicated that in EC, KIAA0101 functions as an oncogene, which is in agreement with nearly all of the literature except one study on hepatic cancer. (iii) Results of aCGH analysis revealed that KIAA0101 knockdown altered gene copy numbers, which could affect gene expression. These genes include MAPK,15 BCL-2,16 and MAL,17 which has been shown in previous studies10, 13, 14 to be altered in EC. In addition, several previously unreported genes (such as LY6K, MFAP5, HOXC9, TNFRSF12A, DHRS2, FST, and GPX3) were also found to be differentially expressed in KIAA0101-knocked-down esophageal cells, which warrants further investigation. This phenomenon suggests that the work on discovering the mechanisms of KIAA0101 function is much more complicated than that first imagined. Although KIAA0101 has only two known motifs, ie, binding to PCNA using PIP box to mediate cell progression18, 19 and binding to ubiquitination enzymes, such as APC/C for degradation of KIAA0101,20 KIAA0101 may regulate a great number of genes involved in the promotion of EC development or progression. Another novelty of the current study is the aCGH experiment that researched the potential mechanisms of KIAA0101, such as NF-κB.21 To the best of our knowledge, this is the first study to demonstrate the genomic copy-number alterations by a single gene alteration in EC cell lines, providing a comprehensive genome-wide gene expression profile and integrative analysis of the genome by KIAA0101. The aCGH screening data also revealed some of genes that are regulated by KIAA0101 that have not been reported previously, such as F3, F7, and F10, which may be related with tumor-associated hypercoagulable state,22 but others do associate with human cancers, such as NGF, which links to perineural invasion (PNI) of cancer in several cancer types, especially in pancreatic cancer.23 This finding suggests that KIAA0101 may be an effective driver gene in Eca-109, but further studies are required to disclose the functions of these genes in EC development and progression and to investigate how KIAA0101 regulates these genes.
In addition, previous studies have indicated that inhibition of KIAA0101 suppressed cell proliferation and invasion of various cancer cells with unidentified molecular mechanisms. In the current study, we demonstrated that KIAA0101 participated in cell mitosis via upregulation of cyclins A and B, which participates in promoting cell from G1 phase to S and G2 phase to M, and may explain the underlying mechanism of KIAA0101-enhaned cancer cell proliferation. Moreover, as KIAA0101 participates in DNA repair after DNA damage, patients with increased KIAA0101 expression could be at a reduced sensitivity to cisplatin-based chemotherapy. Cisplatin is a widely used chemotherapy agent in solid tumors (such as cancers of the liver, lung, esophagus, stomach, and pancreas) and elevated expression of KIAA0101 will facilitate DNA repair but reduce cisplatium antitumor effects, which in turn leads to poor sensitivity of tumor cells to cisplatin in addition to KIAA0101 in cell proliferation. Indeed, a previous study showed that overexpression of KIAA0101 greatly decreased the effect of cisplatin plus doxorubicin in hepatocellular carcinoma.24 Although we did not further explore the underlying molecular mechanisms, this result may provide a novel insight into KIAA0101 for the selection of optimal chemotherapy for EC patients. A further study will pursue such underlying mechanism for effective control of EC.
In conclusion, KIAA0101 is emerging as a meaningful marker for poor prognosis in EC, such as early recurrence and short survival. This study shows that KIAA0101 upregulates cell mitosis via an increase in cyclins A and B, resulting in enhanced cell resistance to cisplatin. These data may provide mechanistic evidence for the reduced chemosensitivity to cisplatin-based therapy seen in EC.
References
van Rensburg SJ . Esophageal cancer, micronutrient malnutrition, and silica fragments. Lancet 1982;2:1098–1099.
Torpy JM, Burke AE, Glass RM . JAMA patient page. Esophageal cancer. JAMA 2010;304:704.
Warbrick E . A functional analysis of PCNA-binding peptides derived from protein sequence, interaction screening and rational design. Oncogene 2006;25:2850–2859.
Yu P, Huang B, Shen M et al. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene 2001;20:484–489.
Simpson F, Lammerts van Bueren K, Butterfield N et al. The PCNA-associated factor KIAA0101/p15(PAF) binds the potential tumor suppressor product p33ING1b. Exp Cell Res 2006;312:73–85.
Petroziello J, Yamane A, Westendorf L et al. Suppression subtractive hybridization and expression profiling identifies a unique set of genes overexpressed in non-small-cell lung cancer. Oncogene 2004;23:7734–7745.
Mizutani K, Onda M, Asaka S et al. Overexpressed in anaplastic thyroid carcinoma-1 (OEATC-1) as a novel gene responsible for anaplastic thyroid carcinoma. Cancer 2005;103:1785–1790.
Guo M, Li J, Wan D et al. KIAA0101 (OEACT-1), an expressionally down-regulated and growth-inhibitory gene in human hepatocellular carcinoma. BMC Cancer 2006;6:109.
Iwase H, Shimada M, Tsuzuki T et al. Concurrent chemoradiotherapy with a novel fluoropyrimidine, S-1, and cisplatin for locally advanced esophageal cancer: long-term results of a phase II trial. Oncology 2013;84:342–349.
Zhu K, Diao D, Dang C et al. Elevated KIAA0101 expression is a marker of recurrence in human gastric cancer. Cancer Sci 2012;104:353–359.
Yang XH, Zou L . Dual functions of DNA replication forks in checkpoint signaling and PCNA ubiquitination. Cell Cycle 2009;8:191–194.
Yuan RH, Jeng YM, Pan HW et al. Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2007;13:5368–5376.
Emanuele MJ, Ciccia A, Elia AE et al. Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc Natl Acad Sci USA 2011;108:9845–9850.
Jain M, Zhang L, Patterson EE et al. KIAA0101 is overexpressed, and promotes growth and invasion in adrenal cancer. PLoS One 2011;6:e26866.
Bao XH, Takaoka M, Hao HF et al. Esophageal cancer exhibits resistance to a novel IGF-1R inhibitor NVP-AEW541 with maintained RAS-MAPK activity. Anticancer Res 2012;32:2827–2834.
Yang HL, Dong YB, Elliott MJ et al. Caspase activation and changes in Bcl-2 family member protein expression associated with E2F-1-mediated apoptosis in human esophageal cancer cells. Clin Cancer Res 2000;6:1579–1589.
Mimori K, Shiraishi T, Mashino K et al. MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene 2003;22:3463–3471.
Maga G, Hubscher U . Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 2003;116:3051–3060.
Moldovan GL, Pfander B, Jentsch S . PCNA, the maestro of the replication fork. Cell 2007;129:665–679.
Povlsen LK, Beli P, Wagner SA et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol 2012;14:1089–1098.
Li K, Ma Q, Shi L et al. NS5ATP9 gene regulated by NF-kappaB signal pathway. Arch Biochem Biophys 2008;479:15–19.
Wang JG, Geddings JE, Aleman MM et al. Tumor-derived tissue factor activates coagulation and enhances thrombosis in a mouse xenograft model of human pancreatic cancer. Blood 2012;119:5543–5552.
Bapat AA, Hostetter G, Von Hoff DD et al. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer 2011;11:695–707.
Liu L, Chen X, Xie S et al. Variant 1 of KIAA0101, overexpressed in hepatocellular carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53 activation. Hepatology 2012;56:1760–1769.
Acknowledgements
The present study was supported by grants from the National Natural Scientific Foundation of China (no. 81001089, no. 30973489, and no. 81000728).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
KIAA0101 is a protein involved in the regulation of DNA repair, cell cycle progression, and migration. Its elevated expression in esophageal cancer is associated with resistance to chemotherapy, recurrence, and poor survival. This paper shows that in esophageal cancer, KIAA0101 enhances cell proliferation, increases resistance to cisplatin treatment, and alters chromosome stability. These findings suggest KIAA0101 as a target for therapy.
Rights and permissions
About this article
Cite this article
Cheng, Y., Li, K., Diao, D. et al. Expression of KIAA0101 protein is associated with poor survival of esophageal cancer patients and resistance to cisplatin treatment in vitro. Lab Invest 93, 1276–1287 (2013). https://doi.org/10.1038/labinvest.2013.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2013.124
Keywords
This article is cited by
-
PCLAF promotes neuroblastoma G1/S cell cycle progression via the E2F1/PTTG1 axis
Cell Death & Disease (2022)
-
KIAA0101, a target gene of miR-429, enhances migration and chemoresistance of epithelial ovarian cancer cells
Cancer Cell International (2016)