Abstract
Cdx2 expression in esophageal stem cells induced by reflux bile acids may be an important factor for development of Barrett's esophagus, whereas Notch signaling is a molecular signaling pathway that plays an important role in the determination of cell differentiation. ATOH1 (a factor associated with Notch signaling) plays an important role in differentiation of stem cells into goblet cells. However, the relationship between the Notch signaling pathway and Cdx2 expression in the development of Barrett's esophagus has not been explored. The aim of this study was to investigate the interrelationship between Notch signaling and Cdx2 in esophageal epithelial cells. The expressions of Cdx2, MUC2, and intracellular signaling molecules related to Notch signaling (Notch1, Hes1, and ATOH1) were examined using real-time polymerase chain reaction (PCR) and immunohistochemical staining with biopsy specimens obtained from esophageal intestinal metaplasia (IM) with goblet cells (IM(+)) and columnar epithelium not accompanied by goblet cells (IM(−)). For in vitro experiments, we employed human esophageal epithelial cell lines (OE33, OE19, and Het-1A). After forced Cdx2 expression by applying a Cdx2 expression vector to the cells, changes in the expressions of Notch1, Hes1, ATOH1, Cdx2, and MUC2 were analyzed by real-time PCR and western blot analysis. Changes in expressions of Notch1, Hes1, ATOH1, Cdx2, and MUC2 in cells were analyzed following stimulation with bile acids in the presence or absence of Cdx2 blocking with Cdx2-siRNA. Suppressed Hes1 and enhanced ATOH1 and MUC2 expressions were identified in IM(+) specimens. Forced expression of Cdx2 in cells suppressed Hes1, and enhanced ATOH1 and MUC2 expressions, whereas bile acids suppressed Hes1, and enhanced ATOH1, Cdx2, and MUC2 expressions. On the other hand, these effects were blocked by siRNA-based Cdx2 downregulation. Enhanced expression of Cdx2 by stimulation with bile acids may induce intestinal differentiation of esophageal columnar cells by interaction with the Notch signaling pathway.
Similar content being viewed by others
Main
Barrett's esophagus is an acquired condition in which normal squamous epithelium of the distal esophagus is replaced by intestinal-type metaplastic columnar epithelium with goblet cells.1 It is considered to be induced by chronic gastroesophageal reflux disease (GERD), and ∼10% of patients with GERD are found to have Barrett's esophagus in an esophagogastroduodenoscopy examination.2 Barrett's esophagus is considered, at least in part, to arise from gastric-type metaplastic epithelium in the esophagus that developed by chronic gastroesophageal reflux. The presence of Barrett's esophagus is one of the predominant risk factors for development of esophageal adenocarcinoma (EAC), and the incidence of EAC derived from Barrett's esophagus has steadily increased during the previous 2 decades in most countries.3, 4 Barrett's esophagus affects ∼2% of the Western population and progresses to EAC in 0.5% of these patients each year.5, 6 Several clinical studies have suggested that reflux of duodenal contents with bile acids contributes to the development of Barrett's esophagus.7, 8, 9, 10 Although there is great interest in the pathogenesis of the condition, little is known regarding the mechanism of cellular metaplasia of Barrett's epithelium.
A number of studies have shown that Cdx2 is one of the most important mediators in the development of Barrett's esophagus.11, 12 We previously reported a two-step mechanism involved in the development of Barrett's epithelium, in which bile acids activate the Cdx2 promoter via nuclear factor-κB (NF-κB) and stimulate the production of Cdx2 protein in esophageal immature keratinocytes, with resulting production of intestinal-type mucin.13 In addition to Cdx2, ATOH1 (a factor associated with Notch signaling) is known to be an important transcriptional activator of the intestinal-specific gene MUC2.14, 15 The Notch signaling pathway is a fundamental molecular signaling system that governs cell-fate decisions such as differentiation, proliferation, and apoptosis in numerous tissue types.16 The function of Notch signaling has been studied in the intestinal epithelium of mice deficient in Hes1, which leads to loss of Notch signaling and an increase in the number of goblet cells and endocrine cells, along with a decrease in absorptive epithelial cells.17 Although analysis of the function of Notch signaling in preservation of esophageal squamous epithelium has not been reported, studies on the effects of Notch signaling in the development of Barrett's esophagus, possibly after loss of homeostasis, have been occasionally presented.18, 19
Recent findings have shown a relationship between the Notch signaling pathway and Cdx2 expression. It was reported that induction of Cdx2 in intestinal epithelium leads to induction of ATOH1, which plays an important role in differentiation of stem cells into goblet cells.20 Furthermore, an analysis of the genes expressed and induced by the Cdx2 gene revealed a significant reduction in the expression of the Hes1 gene following Cdx2 induction.21 Thus, Notch signaling activation leads to Hes1 upregulation and ATOH1 downregulation, along with suppression of stem cell differentiation into goblet cells.15 Therefore, it is plausible that enhanced expression of Cdx2 by chronic inflammatory stimulation (eg, GERD) leads to Hes1 downregulation and ATOH1 upregulation, which in turn enhances the differentiation of stem cells into goblet cells. However, the relationship between the Notch signaling pathway and Cdx2 expression in the development of Barrett's esophagus accompanied by goblet cells has not been reported.
In the present study, we investigated the interaction between the Notch signaling pathway and Cdx2 expression in the development of intestinal-type esophageal metaplasia with goblet cells from columnar metaplasia without goblet. Furthermore, we investigated the complicated mechanisms related to the Notch signaling pathway and Cdx2 expression in the development of esophageal intestinal metaplasia (IM): Barrett's esophagus.
MATERIALS AND METHODS
Patients and Human Esophageal Tissue
A total of 71 patients (43 males, mean 67.7 years old) treated from August 2008 to July 2009 were enrolled in the study, including 44 with columnar-lined esophagus (CLE) and 27 with a normal esophagus. The esophagogastric junction was defined as the proximal margin of the gastric folds. Biopsy samples were precisely taken from CLE specimens, and then snap-frozen in liquid nitrogen. We divided 51 lesions from 44 patients with CLE into two groups based on histopathological findings: (1) lesions of columnar epithelium not accompanied by goblet cells (n=30) and (2) lesions of intestinal columnar epithelium accompanied by goblet cells (n=21). Human tissue specimens were obtained from patients who underwent an endoscopic examination at Shimane University Hospital with a GIF-H260, H260Z, or Q260J endoscope (Olympus Medical Systems, Tokyo, Japan). All endoscopic examinations were performed by well-trained expert endoscopists (TY, NI, SI, and YA), and endoscopic diagnosis was established by consensus of at least three of these four endoscopists. The study protocol was prepared according to the Declaration of Helsinki and approved by the ethics committee of Shimane University, Faculty of Medicine. Written informed consent was obtained from all patients.
Histopathology
Histopathological analysis was performed using tissues sectioned into 5-μm slices, and stained with hematoxylin and eosin (HE) for light microscopy examinations. Biopsy specimens were immediately placed in a 10% buffered formalin solution, routinely processed, and embedded in paraffin, with nine sequential sections obtained. Two sections were stained with HE and periodic acid-Schiff (PAS) Alcian blue staining, and a histological diagnosis of Barrett's esophagus was made when specialized columnar epithelium containing goblet cells with acid mucin was identified in the biopsy specimen by PAS Alcian blue staining.22 The remaining seven sequential sections were used for immunohistochemical examinations, as described below. The mucin phenotype was determined following histological diagnosis of Barrett's epithelium with the biopsy specimens. Antibodies against human gastric mucin (HGM) (Novocastra, Newcastle, UK) and MUC2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used for phenotyping of Barrett's esophagus, as previously described.23 When Barrett's epithelial cells were more dominantly stained by the anti-MUC2 antibody, the case was diagnosed as Barrett's esophagus with intestinal predominant mucin phenotype.
Immunohistochemistry
Immunolabeling was performed using formalin-fixed, paraffin-embedded blocks (5 μm thick). Freshly cut sections were deparaffinized in xylane and rehydrated through sequential graded ethanol steps. The tissue sections were incubated with primary antibodies, followed by incubation with secondary biotinylated antibodies (DAKO, Carpinteria, CA, USA). Bound antibodies were detected using an avidin-biotin peroxidase method (ABC Elite Kit; Vector Laboratories, Burlingame, CA, USA). The sections were then counterstained with HE. The antibodies and dilutions used are summarized in Table 1. Mounted slides were examined by light microscopy and immunoreactivity was assessed by four independent observers (YT, NI, GU, and YA) using a three-grade system, in which 0 denoted absence of staining; 1 denoted minimal and variable staining; and 2 denoted obvious, uniform, and intense staining. Only specimens with grade 2 immunoreactivity were considered positive.
Cell Culture and Treatments
Three cell lines, including OE33 (a human esophageal adenocarcinoma cell line; European Collection of Cell Cultures (ECACC), Salisbury, Wiltshire, UK), OE19 (a human cell line established from adenocarcinoma obtained from gastric cardia/esophageal gastric junction; ECACC), and Het-1A (a human esophageal squamous epithelial cell line immortalized by viral SV40 transfection; American Type Culture Collection (ATCC), Manassas, VA, USA) were used in this study. OE33 cells are poorly differentiated and OE19 cells are moderately differentiated EAC cells.24 All cells were maintained in monolayer culture at 37 °C in a humidified incubator containing 5% carbon dioxide (CO2). OE33 cells and OE19 cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) medium supplemented with 2 mM glutamine, 10% fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 μg/ml streptomycin (all from Invitrogen). Het-1A cells were cultured in BEBM (Lonza, Walkersville, MD, USA) medium supplemented with 100 U/ml penicillin G and 100 μg/ml streptomycin (all from Invitrogen). Subconfluent cells were split and subcultured on plastic dishes until 70% confluent, and then serum-starved 24 h before the experiments. All cell culture experiments were performed in triplicate to ensure reproducibility. After reaching ∼70% confluence, the cells were incubated with increasing concentrations of deoxycholic acid (DCA) or cholic acid (CA) (all from Sigma Chemical, St Louis, MO, USA; 50, 100, and 200 μM for each cell line) at neutral pH for up to 12 h, or with an increasing concentration of the γ-secretase inhibitor N-(N-(3,5-difluorophenacetyl-L-aranyl)-S-phenylglicine)-t-butyl ester (DAPT) (Calbiochem, Darmstadt, Germany; 5, 10, and 20 μM for each cell line) for up to 48 h. Inhibition of Notch signaling was achieved by addition of DAPT. For the experimental groups at neutral pH, the media were titrated to the corresponding pH before addition of the other components and pH was verified again thereafter. The cells were then incubated for the indicated time periods. RNAs or proteins were extracted as described below. Incubation time, DCA, CA, DAPT, and concentrations were chosen according to the criteria described elsewhere.18, 25, 26, 27
RNA Extraction and Complementary DNA (cDNA) Syntheses
Total RNA was extracted using a single-step guanidium thiocyanate-phenol-chloroform method (Isogen; Nippon Gene, Tokyo, Japan), according to the manufacturer's instructions. Total RNA concentrations were determined by spectrophotometric quantification at 260 nm using Nanodrop (type ND-1000; NanoDrop Technologies, Wilmington, DE, USA) and the integrity of the bands was verified visually by agarose gel electrophoresis. For real-time reverse-transcription polymerase chain reaction (PCR) assays, cDNA was synthesized from 0.5 μg total RNA using an AffintyScript™ QPCR cDNA Synthesis Kit (Stratagene, La Jolla, CA, USA), according to the manufacturer's protocol, and the final reaction was diluted fivefold in RNase-free water.
Real-Time PCR
For real-time PCR, 2 μl of cDNA and primers were mixed with POWER SYBR- Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in a final reaction volume of 20 μl, then assayed using a StepOnePlus™ Real Time PCR System (Applied Biosystems) as directed by the manufacturer. We used primer sequences for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Notch1, Hes1, ATOH1, Cdx2, and MUC2. The GAPDH subunit was used as the normalization control. Notch1, Hes1, ATOH1, Cdx2, and MUC2 messenger RNAs (mRNAs) were quantitated using real-time PCR technology. The primer sequences used and amplicon size are summarized in Table 2. Real-time PCR products were subjected to melting curve analysis. Agarose gel electrophoresis showed the expected band. Data are expressed as the ratio of product copies/μl to copies/μl of the housekeeping gene GAPDH from the same RNA (respective complementary DNA) sample and subjected to PCR. Results are expressed as the mean±s.e.m. of data collected from three rounds of assays. Each assay was performed in triplicate experiments.
Protein Extraction for Western Blot Analysis
Cells were washed with phosphate-buffered saline (PBS) twice and lysed for 5 min on ice with RIPA Buffer (25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and a Protease Inhibitor Cocktail Kit (protease inhibitor cocktail and 0.5 M ethylenediaminetetraacetic acid (EDTA) solution; all from Pierce Biotechnology, Rockford, IL, USA). Cell lysates were subjected to centrifugation at 14 000 g for 15 min at 4 °C. The supernatants were collected and protein concentrations determined using a bicinchoninic acid (BCA) protein assay reagent kit (Pierce Biotechnology) according to the manufacturer's instruction manual. Absorbance at 545 nm was measured using Nanodrop (NanoDrop Technologies). Bovine serum albumin (BSA) served as the standard. The lysates were diluted 1:1 in Electrophoresis Sample Buffer (glycerol, 2-mercaptoethanol, SDS, 1.0 M Tris-HCl (pH 6.7), and bromophenol blue; Santa Cruz Biotechnology) and incubated for 5 min at 95 °C.
Western Blot Analysis
Samples (20 μg/lane) were analyzed using 6 or 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli.28 Proteins were electro-transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Hybond-P PVDF membrane; GE Healthcare, Buckinghamshire, UK) in transfer buffer containing 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 20% methanol (80 V for 6 h at room temperature). After incubation in blocking buffer (either 5% non-fat milk or 5% BSA in Tris-buffered saline supplemented with 0.1% Tween 20 (TBST)), the blots were incubated overnight at 4 °C with the appropriate primary antibodies. Blots were then washed with TBST and incubated for 1 h at room temperature in 1:3000 horseradish peroxidase-conjugated secondary antibodies (DAKO). After a final wash with TBST, bound antibodies were visualized using a chemiluminescent substance (Amersham ECL Plus Western Blotting Reagents Components/Solution B; GE Healthcare) and exposed to New Amersham Hyperfilm ECL (GE Healthcare). The mean results of three separate assays are shown and each assay was performed in triplicate. The primary antibodies and dilutions used are summarized in Table 1.
Expression Plasmid and Transient Transfection
The cDNA encoding full-length mouse Cdx2 was amplified by PCR using the following primers: 5′-AGCTTTCAGACTCCGGAGCCTCAGC-3′ and 5′-GTTGGACCCAGCTGGGCAAGAAAT-3′ (NCBI NM_007673). Amplified DNA samples were cloned into a pcDNATM 5/FRT/V5-His TOPO vector (Invitrogen) and the plasmid clones were sequenced to confirm the constructs. OE33, OE19, and Het-1A cells were separately grown in six-well dishes, and then transfected with the expression vector in each well, using LipofectamineTM 2000 Transfection Reagent (Invitrogen). Vector DNA without Cdx2 sequences was used as a negative control. After incubation at 37 °C in a humidified incubator containing 5% CO2 for 6 h, 2 ml of normal growth medium was exchanged. Samples were then prepared and analyzed using real-time PCR and western blot analysis as described previously.
Small Interfering RNA (siRNA) Knockdown of Cdx2 Expression
The siRNA, a specific double-stranded 20–25-nucleotide RNA sequence homologous to the target gene, was used to silence Cdx2 expression. The siRNA oligonucleotide for human Cdx2 was purchased from Santa Cruz Biotechnology (sc-43680). Inhibition of RNA or protein expression was assessed after transient transfection of OE33 and OE19 cells with siRNA. In brief, cells were grown in six-well dishes and transiently transfected with 10 nM of pooled siRNAs using 0.4 μl/ml Lipofectamine 2000 Transfection Reagent (Invitrogen) in a total transfection volume of 2 ml of Dulbecco's modified Eagle's medium containing 10% FBS. After incubation at 37 °C in a humidified incubator containing 5% CO2 for 6 h, 2 ml of normal growth medium was exchanged. Samples were then prepared and analyzed by real-time PCR and western blot analyses as described previously.
Statistical Analysis
All data were obtained from at least three independent experiments and are expressed as the mean±s.e.m., unless otherwise indicated. Multiple comparisons were performed with ANOVA, followed by Dunnett's test. Differences with a P-value of <0.05 were considered to be statistically significant.
RESULTS
Immunohistochemical Examination of Human Esophageal Tissue Specimens
To elucidate the parameters of Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2, we used immunohistochemistry to investigate sections of human esophageal IM with goblet cells (IM(+)) and columnar epithelia not accompanied by goblet cells (IM(−)). The numbers and clinical characteristics of patients included in this study are shown in Table 3. No significant difference was found between clinical characteristics of IM(−) and that of IM(+) samples. Cdx2- and ATOH1-positive cells with nuclear staining were observed in the IM(+) samples. Nuclei in most goblet cells were labeled with the antibodies to Cdx2 and ATOH1, although goblet cells occasionally lacked nuclear staining. In contrast, the IM(−) samples exhibited weaker and patchy staining, or no staining (Figure 1a). Similarly, immunohistochemical findings of biopsy specimens demonstrated the expression of MUC2 in IM(+) samples, whereas there was little expression in IM(−) samples (data not shown). MUC2 positively stained all of 21 IM(+) samples. Cdx2 and ATOH1 were found in 18 of 21 IM(+) samples with similar frequency and immunoreactivity (Table 4). Next, we assessed the expressions of Notch1 and Hes1 by immunohistochemistry. Notch1 expression in IM(+) and IM(−) samples was typically localized in the cell membrane and cytoplasm of esophageal epithelial cells with no difference in Notch1 immunoreactivity between IM(+) and IM(−) samples. Hes1 expression in IM(−) samples was clearly found in the nucleus as well as in cytoplasm, whereas the IM(+) samples exhibited weaker and patchy staining (Figure 1a).
Expressions of Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2 in human esophageal intestinal metaplasia (IM) with goblet cells (IM(+)) and columnar epithelium without goblet cells (IM(−)). (a) Immunohistochemical detection of Cdx2, ATOH1, Notch1, and Hes1 in biopsy specimens taken from IM(+) and IM(−) (original magnification × 400). (b) Expressions of Notch1, Hes1, ATOH1, Cdx2, and MUC2 mRNA in Barrett's epithelium with IM(+) and IM(−). Results are expressed as the mean±s.e.m. of three experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from IM (−). *P<0.05, statistically significant difference vs IM(−). **P<0.01, statistically significant difference vs IM(−). NS indicates that the comparison was not significant.
Expressions of ATOH1 and Cdx2 mRNA in Human Esophageal Columnar Epithelia with and without Goblet Cells Positing IM
We also determined Notch1, Hes1, ATOH1, Cdx2, and MUC2 mRNA expression levels in endoscopic biopsy specimens from IM(+) and IM(−) esophageal columnar metaplasia. ATOH1, Cdx2, and MUC2 mRNA expression levels in IM(+) samples were significantly greater than those in IM(−) samples, whereas the Hes1 mRNA expression level in IM(+) samples was significantly lower than in IM(−) samples, with no significant difference between IM(+) and IM(−) samples in Notch1 mRNA expression levels (Figure 1b).
Effects of Notch Signaling Pathway-Specific Inhibitor on Cdx2 and MUC2 Expressions in Human Esophageal Cells
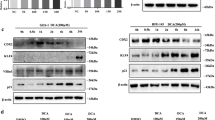
DAPT is known as a specific γ-secretase inhibitor of the Notch signaling pathway.29, 30, 31 To determine whether DAPT augments Cdx2 and MUC2 mRNA expressions, we investigated its effects on Cdx2 and MUC2 mRNA expressions using OE33, OE19, and Het-1A cells, and found that DAPT augmented those expressions in concentration- and time-dependent manners. Similarly, effective inhibition was confirmed by Hes1 and ATOH1 mRNA expressions. ATOH1 mRNA expression was upregulated by DAPT, whereas that of Hes1 was decreased, in concentration- and time-dependent manners (Figures 2a–c). We also investigated the effects of DAPT on Cdx2 and MUC2 protein expressions in OE33 and OE19 cells, and found that it augmented Cdx2 and MUC2 protein expressions in a concentration-dependent manner. Effective inhibition was confirmed by expression of the intracellular-γ-secretase cleaved Notch1 fragment using a specific antibody that recognizes only the cleaved fragment, as well as by Hes1 and ATOH1 protein expressions. DAPT decreased cleaved Notch1 protein expression in OE33 and OE19 cells, whereas Hes1 protein was significantly decreased and ATOH1 increased (Figures 2d and e).
Effects of DAPT on Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2 expressions in OE33, OE19, and Het-1A cells. Hes1 mRNA levels were downregulated, whereas ATOH1, Cdx2, and MUC2 mRNA levels were upregulated by the γ-secretase inhibitor DAPT in concentration- and time-dependent manners. (a) OE33, (b) OE19, and (c) Het-1A cells were exposed to DAPT for 12, 24, and 48 h at concentrations of 0, 5, 10, and 20 μM. Expressions of the indicated genes were quantitated by real-time PCR. The results are expressed as the mean±s.e.m. of four experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from the control. *P<0.05, statistically significant difference vs control. **P<0.01, statistically significant difference vs control. NS indicates that the comparison was not significant (n=4 for each group). Effects of DAPT on cleaved Notch1, Hes1, ATOH1, Cdx2, and MUC2 protein expressions in (d) OE33 and (e) OE19 cells. Cells were stimulated with various concentration of DAPT or the vehicle alone for 24 h, and then proteins were extracted and subjected to western blot analysis for cleaved Notch1, Hes1, ATOH1, Cdx2, MUC2, and β-actin. Blots shown are representative of three separate experiments.
Effects of Cdx2 Overexpression on Hes1, ATOH1, and MUC2 Expressions in Human Esophageal Cells
To investigate whether Cdx2 overexpression influences ATOH1 mRNA expression in esophageal epithelial cells, we transfected a Cdx2 expression vector into OE33, OE19, and Het-1A cells, and determined the levels of ATOH1 and Hes1 mRNA expressions. After 48 h, cells transfected with the Cdx2 expression construct induced ATOH1 mRNA expression, whereas Hes1 mRNA expression was suppressed. In addition, cells transfected with the Cdx2 expression vector showed induced expression of intestine-specific MUC2 mRNA, and there was no difference between Cdx2-transfected and empty vector-transfected cells with regard to Notch1 mRNA expression level. Het-1A cells responded in the same fashion as OE33 and OE19 cells to Cdx2 overexpression (Figures 3a–c). Next, ATOH1 protein expression following stimulation with a Cdx2 expression vector was evaluated in OE33 and OE19 cells using western blot analysis. Cells transfected with the Cdx2 expression construct induced ATOH1 and suppressed Hes1 protein expression. Similarly, transfection of the Cdx2 expression vector into those cells induced MUC2 protein expression, with no difference between Cdx2-transfected and empty vector-transfected cells with regard to Notch1 protein expression level (Figures 3d and e).
Effects of Cdx2 overexpression on Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2 expressions in OE33, OE19, and Het-1A cells. RNA was extracted 48 h after transfection in (a) OE33, (b) OE19, and (c) Het-1A cells, reverse transcribed to cDNA, and subjected to real-time PCR. Results are expressed as the mean±s.e.m. of four experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from an empty vector. **P<0.01, statistically significant difference vs empty vector. NS indicates that the comparison was not significant (n=4 for each group). (d) OE33 and (e) OE19 cells were transfected with either an empty vector or Cdx2 expression vector. Proteins were extracted 48 h after transfection in OE33 and OE19 cells, and then subjected to western blot analysis for Notch1, Hes1, ATOH1, Cdx2, MUC2, and β-actin. Blots shown are representative of three separate experiments.
Effects of Bile Acids on Hes1, ATOH1, Cdx2, and MUC2 Expressions in Human Esophageal Cells
Bile acids are known to play an important role in the induction of Cdx2 expression and we examined their effects on the Notch signaling pathway. To determine their influence, we investigated the effects of bile acids on Hes1, ATOH1, and Cdx2 mRNA expressions using OE33, OE19, and Het-1A cells. Our results showed that bile acids augmented ATOH1 and Cdx2 mRNA expressions in concentration- and time-dependent manners in all cell lines, whereas MUC2 mRNA expression was consistently augmented by bile acids in all cell lines. On the other hand, Hes1 mRNA expression was suppressed by stimulation with bile acids in concentration- and time-dependent manners in all cell lines (Figures 4a–c and f–h). Therefore, blockade of the Notch signaling pathway by bile acids was followed by inhibition of Hes1 and activation of ATOH1 at mRNA expression levels. Ordinarily, ATOH1 synthesis is inhibited by Hes1. Thus, we also investigated the effects of bile acids on cleaved Notch1, Hes1, ATOH1, Cdx2, and MUC2 protein expressions in all cell lines, and found that ATOH1, Cdx2, and MUC2 protein expressions were augmented in a concentration-dependent manner, whereas Hes1 protein expression was decreased in a concentration-dependent manner, with no difference in cleaved Notch1 protein expression levels (Figures 4d and e). These findings indicate that stimulation with bile acids decreased the Notch signaling pathway, thereby increasing ATOH1 and resulting in Cdx2 and MUC2 expressions in all cell lines.
Effects of deoxycholic acid (DCA) on Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2 expressions in OE33, OE19, and Het-1A cells. Hes1 mRNA levels were downregulated, whereas those of ATOH1, Cdx2, and MUC2 were upregulated by DCA in concentration- and time-dependent manners. (a) OE33, (b) OE19, and (c) Het-1A cells were exposed for 2, 4, 8, and 12 h to DCA at concentrations of 0, 50, 100, and 200 μM. Expressions of the indicated genes were quantitated by real-time PCR. Results are expressed as the mean±s.e.m. of four experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from the control. *P<0.05, statistically significant difference vs control. **P<0.01, statistically significant difference vs control. NS indicates that the comparison was not significant (n=4 for each group). Effects of DCA on cleaved Notch1, Hes1, ATOH1, Cdx2, and MUC2 protein expressions in (d) OE33 and (e) OE19 cells. Cells were stimulated with various concentrations of DCA or the vehicle alone for 8 h, and then proteins were extracted and subjected to western blot analysis for cleaved Notch1, Hes1, ATOH1, Cdx2, MUC2, and β-actin. Blots shown are representative of three separate experiments. (f) OE33, (g) OE19, and (h) Het-1A cells were exposed for 2, 4, 8, and 12 h to CA at concentrations of 0, 50, 100, and 200 μM. Expressions of the indicated genes were quantitated by real-time PCR. Results are expressed as the mean±s.e.m. of four experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from the control. *P<0.05, statistically significant difference vs control. **P<0.01, statistically significant difference vs control. NS indicates that the comparison was not significant (n=4 for each group).
Effects of Cdx2-Specific-siRNA on Hes1, ATOH1, and MUC2 Expressions in Human Esophageal Cells
To determine whether ATOH1 induction by bile acids occurs via Cdx2 activation, we employed the siRNA approach to inhibit endogenous Cdx2 in OE33 and OE19 cells. In Cdx2 siRNA-transfected samples, both Cdx2 mRNA and protein expressions were significantly blocked (Figures 5a, d, and e). Furthermore, ATOH1 and MUC2 mRNA expressions were significantly decreased, whereas Hes1 mRNA expression was augmented in cells transfected with specific Cdx2 siRNA as compared with the control nonspecific siRNA-transfected cells following bile acid treatment (Figures 5b and c). Cdx2 protein expression was also clearly knocked down by treatment with Cdx2-specific siRNA, completely abrogating any increase in Cdx2 expression upon treatment with bile acids. Notch1 expression was not affected by treatment with the siRNA targeted to Cdx2 (data not shown). In addition, knockdown of Cdx2 significantly downregulated the expressions of both ATOH1 and MUC2, whereas Hes1 expression was upregulated in cells treated and not treated with bile acids (Figures 5d and e).
Effects of Cdx2-specific siRNA on Notch signaling (Notch1, Hes1, and ATOH1), Cdx2, and MUC2 expressions in OE33 and OE19 cells. (a) siRNA-mediated knockdown of Cdx2 downregulated Cdx2 mRNA expression in OE33 and OE19 cells. Cells were transfected with either Cdx2-specific siRNA or control nonspecific siRNA and cultured for 48 h, and then RNA was extracted and subjected to real-time PCR. Results are expressed as the mean±s.e.m. of four experiments. Data were normalized to the expression level of GAPDH. A value of 1 for this ratio was assigned arbitrarily to the data obtained from the control nonspecific siRNA. **P<0.01, statistically significant difference (n=4 for each group). (b–e) At 48 h after transfection with each siRNA, OE33 (b) and OE19 (c) cells were stimulated with DCA (200 μM) or the vehicle alone for 8 h, and then RNA was extracted and subjected to real-time PCR. Data are expressed as the n-fold increase in transcripts in DCA-stimulated samples over that in the vehicle-treated samples. Results are expressed as the mean±s.e.m. of four experiments. *P<0.05 **P<0.01, statistically significant difference vs control siRNA-transfected samples with DCA treatment. NS indicates that the comparison was not significant (n=4 for each group). In the same condition, proteins were also extracted for western blot analysis (d, e). Blots shown are representative of three separate experiments. (f–j) At 48 h after transfection with each siRNA, OE33 (f) and OE19 (g) cells were stimulated with DAPT (20 μM) or the vehicle alone for 24 h, and then RNA was extracted and subjected to real-time PCR. Data are expressed as the n-fold increase in transcripts in DAPT-stimulated samples over that in the vehicle-treated samples **P<0.01, statistically significant difference vs control siRNA-transfected samples with DAPT (n=4 for each group). Expression of the indicated ATOH1 mRNA (h) in OE33 and OE19 cells was quantitated by real-time PCR. Results are expressed as the mean±s.e.m. of four experiments. **P<0.01, statistically significant difference vs control siRNA-transfected samples without DAPT treatment. NS indicates that the comparison was not significant (n=4 for each group). In the same condition, proteins were also extracted for western blot analysis (i, j).
To confirm whether MUC2 induction by DAPT occurs via Cdx2 activation, we employed treatment with DAPT and Cdx2-specific siRNA in OE33 and OE19 cells. Real-time PCR analysis showed knockdown of Cdx2 mRNA expression in cells treated with Cdx2-specific siRNA (Figure 5a). In addition, Hes1 mRNA expression was decreased, whereas ATOH1 mRNA expression was upregulated in DAPT-treated cells. Furthermore, MUC2 mRNA expression was significantly decreased in cells transfected with specific Cdx2 siRNA as compared with the control nonspecific siRNA-transfected cells following DAPT treatment (Figures 5f–h). Western blot analysis revealed that Cdx2 protein expression was also clearly knocked down by treatment with Cdx2-specific siRNA, completely abrogating any increase in MUC2 expression upon treatment with DAPT (Figures 5i and j). In contrast, ATOH1 expression was not affected by treatment with the siRNA targeted to Cdx2. Knockdown of Cdx2 significantly downregulated the expression of MUC2 in cells treated with the vehicle alone. However, upon knockdown of Cdx2 expression, a significant downregulation of MUC2 expression was also observed in cells treated with DAPT. Knockdown of Cdx2 completely abrogated the upregulation of MUC2 expression by DAPT, suggesting that induction of the goblet cell phenotype in Notch-inactivated esophageal epithelial cells is dependent upon upregulation of Cdx2 expression.
DISCUSSION
The present study is the first to demonstrate that the transcriptional network related to the intestine-specific homeobox gene Cdx2 as well as the basic helix-loop-helix transcription factor ATOH1 have important roles in the development of Barrett's epithelium. ATOH1 has been reported to be expressed in epithelial cells of the gut and skin, as well as in other organs.17, 32, 33, 34, 35 However, it has not been fully revealed whether ATOH1 is expressed in the normal esophagus or that with various types of columnar metaplasia. Cdx2 is reported to be expressed in Barrett's epithelium and a number of studies have found that it is a key mediator in the development of Barrett's esophagus.11, 21, 25, 35 Previously, we showed that bile acids activated the Cdx2 promoter via NF-κB and stimulated the production of Cdx2 protein in esophageal keratinocytes, which is an important step in the development of Barrett's epithelium.13 In addition to Cdx2, ATOH1 is also reported to be expressed in Barrett's metaplasia.18, 19 However, the roles of ATOH1 in the development of Barrett's esophagus remain to be reported. A previous study found that overexpression of Cdx2 induced IM in the stomachs of transgenic mouse and suggested that ATOH1 has a role in the development of IM in that mouse model.20 Also, Morrow et al18 suggested that ATOH1 is an important molecular mediator of Barrett's metaplasia and that bile acids stimulate the expression of Cdx2 via ATOH1 expression. Therefore, it is considered that ATOH1 and Cdx2 may have similar roles, and interact with each other in the development of Barrett's esophagus.
In the present study, we found that ATOH1 expression was upregulated in human esophageal IM with goblet cells, but not in columnar epithelium without goblet cells, whereas Hes1 expression was suppressed in esophageal columnar epithelium with IM. Therefore, these results are consistent with previous reports from other groups showing that ATOH1 expression is upregulated in IM of the gut and skin, as well as in other organs.17, 31, 32, 33, 34 Although different types of cells are present in biopsy samples, our results from human tissues suggest that ATOH1 expression is found in Barrett's epithelium.
The resemblance of metaplastic Barrett's epithelium to colonic epithelium prompted us to apply our insights gained in the study of intestinal epithelium to investigate Barrett's epithelium. Notch signaling plays a dominant role in cell-fate decisions in normal colonic epithelium and, when blocked, proliferative epithelial cells instantaneously convert into goblet cells.15 The same phenomenon occurs in adenomas of the intestine upon inhibition of the Notch signaling pathway.36 In the present study, we examined whether the inhibition of Notch signaling in esophageal epithelial cells would trigger a similar transdifferentiation to intestinal goblet-type columnar epithelial cells. In γ-secretase inhibitor DAPT-treated Barrett's adenocarcinoma cell lines, suppressed Hes1 and forced ATOH1 expression by inhibition of Notch signaling induced Cdx2 and MUC2 expressions. The previous report by Leow et al15 showed that forced ATOH1 expression in colon cancer cells results in increased MUC2 expression. Moreover, ATOH1 rodent homolog Math1 is required for the development of secretory cells, including goblet cells, in embryonic mouse intestines.14 Consistent with previous reports, overexpression of ATOH1 by DAPT induced MUC2 mRNA and protein expressions in the present esophageal epithelial cells. However, ATOH1 is a weak inducer of MUC2 as compared with Cdx2, as activation of MUC2 by a Cdx2 expression vector caused an ∼200-fold increase, whereas that induced by overexpression of ATOH1 with DAPT was ∼2-fold. Furthermore, in Cdx2-specific siRNA-treated cells, the γ-secretase inhibitor DAPT did not augment the expression of MUC2 (Figures 5f–j). Therefore, MUC2 expression observed in ATOH1-activated cells is considered to be indirect via the expression of Cdx2. Cdx2 is a transcriptional activator of a number of intestine-specific genes, including MUC2, sucrose-isomaltase, guanylyl cyclase C, and intestinal alkaline phosphatase (IAP).37, 38, 39, 40, 41 Furthermore, ATOH1 also contributes to induction of the inter-regulation network with Cdx2 and indirectly stimulates cellular transdifferentiation into IM.
As a next step, we investigated the inter-regulation mechanism between Cdx2 and ATOH1 in detail. In the present study, we found that forced expression of Cdx2 in the studied cells induced ATOH1 and MUC2 expressions, whereas it suppressed Hes1 expression (Figure 3). Moreover, MUC2 upregulation correlated very well with Cdx2 upregulation. Simultaneous upregulation of both Cdx2 and MUC2 after transfection of a Cdx2 expression vector into Barrett's adenocarcinoma cells is consistent with previous findings from other groups showing that Cdx2 is directly involved in transcriptional regulation of the MUC2 gene in gastric and colon cancer cells.39, 40, 41 Furthermore, the present results raise the possibility that forced expression of Cdx2 decreases the Notch signaling pathway, thereby increasing ATOH1 expression via Hes1 suppression in esophageal epithelial cells. However, the mechanisms by which ATOH1 induction occurs via Cdx2 activation have not been fully revealed.
We also investigated whether bile acids induce ATOH1 and Cdx2 expressions in Barrett's adenocarcinoma cells in vitro. In the present experiments, we found that bile acids augmented ATOH1 and Cdx2 expressions in concentration- and time-dependent manners, whereas that of Hes1 was decreased in the same manner, with no difference in active Notch signaling (cleaved Notch1) expression levels. Furthermore, our results indicate that stimulation with bile acids decreased the Notch signaling pathway, thereby increasing ATOH1 expression via Hes1 suppression and Cdx2 expression in esophageal epithelial cells. However, the mechanisms by which ATOH1 induction occurs following stimulation with bile acids and Cdx2 activation are unknown. Thus, we sought to determine whether bile acids induce ATOH1 and MUC2 expressions via Cdx2 activation in the present cell lines. In Cdx2-specific siRNA-treated cells, bile acids did not change the expression of either ATOH1 or MUC2. Our results also indicate that knockdown of Cdx2 expression abrogated the upregulation of MUC2 expression by bile acids and suggest that stimulation with bile acids induced ATOH1 expression via Cdx2 activation in esophageal epithelial cells. Therefore, it is considered that upregulation of MUC2 expression by bile acids occurs mainly via Cdx2 activation, and our findings also suggest an inter-regulation mechanism between Cdx2 and ATOH1 that contributes to goblet cell formation in the development of Barrett's epithelium. Taken together with our previous findings,13 these results indicate that overexpression of the transcription factors Cdx2 and ATOH1 induced by bile acids may change the phenotype of esophageal stem cells into columnar cells (Figure 6). Recent studies have revealed that the Notch signaling pathway is important for esophageal epithelial homeostasis42 and development of Barrett's adenocarcinoma.43 Further research is required for elucidation of the role of the Notch signaling pathway in the development of Barrett's epithelium.
There are several limitations in this study. One of them is that the study was done only on Barrett's adenocarcinoma cell lines (OE33 and OE19) and esophageal squamous epithelial cell line (Het-1A cells). Because cancer cell lines can have other genetic alterations or activated signaling pathways, it is unclear that the metaplasia model they propose is relevant to Barrett's esophagus without dysplasia (intestinal esophageal epithelial metaplasia). Importantly, however, Het-1A cells responded in the same fashion as OE33 and OE19 cells in this study. Our results should be confirmed using Barrett's cell lines (CP-A, BAR-T, and so on) in the near future.
In conclusion, we found that induction of ATOH1 expression via Hes1 suppression in esophageal epithelial cells in response to bile acids has important functions in the induction of metaplastic changes during Barrett's epithelium development. In addition, our results revealed that the transcriptional network related to the Notch signaling pathway and Cdx2 also has important roles in that development.
References
Jankowski JA, Harrison RF, Perry I, et al. Barrett's metaplasia. Lancet 2000;356:2079–2085.
Paulson TG, Reid J . Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer cell 2004;6:11–16.
Cameron AJ, Zinsmeister AR, Ballard DJ, et al. Prevalence of columnar-lined (Barrett's) esophagus. Gastroenterology 1990;99:918–922.
Blot WJ, Devessa SS, Kneller RW, et al. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA 1991;265:1287–1289.
Ronkainen J, Aro P, Stroskrubb T, et al. Prevalence of Barrett's esophagus in the general population: an endoscopic study. Gastroenterology 2005;129:1825–1831.
van Soest EM, Dieleman JP, Siersema PD, et al. Increasing incidence of Barrett's oesophagus in the general population. Gut 2005;54:1062–1066.
Gillen P, Keeling P, Byme PJ, et al. Implication of duodenogastric reflux in the pathogenesis of Barrett's oesophagus. Br J Surg 1988;75:540–543.
Iftikhar SY, Lendrum K, Ledingham S, et al. Bile reflux in columnar lined Barrett's oesophagus. Ann R Coll Surg Engl 1993;75:411–416.
Nehra D, Howell P, Williams CP, et al. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut 1999;44:598–602.
Stein HJ, Feussner H, Kauer W, et al. Alkaline gastroesophageal reflux: assessment by ambulatory esophageal aspiration and pH monitoring. Am J Surg 1994;167:163–168.
Phillips RW, Frierson Jr HF, Moskaluk CA . Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. Am J Surg Pathol 2003;27:1442–1447.
Moons LM, Bax DA, Kuipers EJ, et al. The homeodomain protein CDX2 is an early marker of Barrett's oesophagus. J Clin Pathol 2004;57:1063–1068.
Kazumori H, Ishihara S, Rumi MA, et al. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut 2006;55:16–25.
Yang Q, Bermingham NA, Finegold MJ, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 2001;294:2155–2156.
Leow CC, Romero MS, Ross S, et al. down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res 2004;64:6050–6057.
Artavanis-Tsakonas S, Rand MD, Lake RJ . Notch Signaling: cell fate control and signal integration in development. Science 1999;284:770–776.
Jensen J, Pedersen EE, Galante P, et al. Control of endodermal endocrine development by Hes-1. Nat Genet 2000;24:36–44.
Morrow DJ, Avissar NE, Toia L, et al. Pathogenesis of Barrett's esophagus: bile acids inhibit the Notch signaling pathway with induction of CDX2 gene expression in human esophageal cells. Surgery 2009;146:714–722.
Menke V, van Es JH, de Lau W, et al. Conversion of metaplastic Barrett's epithelium into post-mitotic goblet cells by γ-secretase inhibition. Dis Model Mech 2010;3:104–110.
Mutoh H, Sakamoto H, Hayakawa H, et al. The intestine-specific homeobox gene Cdx2 induces expression of the basic helix-loop-helix transcription factor Math1. Differentiation 2006;74:313–321.
Liu T, Zhang X, So CK, et al. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis 2007;28:488–496.
Sampliner RE . Practice guidelines on the diagnosis, surveillance, and therapy of Barrett's esophagus. The Practice Parameter Committee of the American College of Gastroenterology. Am J Gastroenterol 1998;93:1028–1032.
Amano Y, Kushiyama Y, Yuki T, et al. Prevalence of and risk factors for Barrett's esophagus with intestinal predominant mucin phenotype. Scand J Gastroenterol 2006;41:873–879.
de Both NJ, Wijnhoven BP, Sleddens HF, et al. Establishment of cell lines from adenocarcinomas of the esophagus or gastric cardia growing in vivo and in vitro. Virchows Arch 2001;438:451–456.
Hu Y, Williams VA, Gellersen O, et al. The pathogenesis of Barrett's esophagus: secondary bile acids upregulate intestinal differentiation factor CDX2 expression in esophageal cells. J Gastrointest Surg 2007;11:827–834.
Avissar NE, Toia L, Hu Y, et al. Bile acid alone, or in combination with acid, induces CDX2 expression through activation of the epidermal growth factor receptor (EGFR). J Gastrointest Surg 2009;13:212–222.
Debruyne PR, Witek M, Gong L, et al. Bile acids induce ectopic expression of intestinal guanylyl cyclase C through nuclear factor-kappa B and Cdx2 in human esophageal cells. Gastroenterology 2006;130:1191–1206.
Laemmli UK . Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–685.
Dovey HF, John V, Anderson JP, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem 2001;76:173–181.
Geling A, Steiner H, Willem M, et al. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep 2002;3:688–694.
Ben-Arie N, Bellen HU, Armstrong DL, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature 1997;390:169–172.
Bermingham NA, Hassan BA, Price SD, et al. Math1: an essential gene for the generation of inner ear hair cells. Science 1999;284:1837–1841.
Gowan K, Helms AW, Hunsaker TL, et al. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal intemeurons. Neuron 2001;31:219–232.
Leonard JH, Cook AL, Van Gele M, et al. Proneural and proneuroendocrine transcription factor expression in cutaneous mechanoreceptor (Merkel) cells and Merkel cell carcinoma. Int J Cancer 2002;101:103–110.
Eda A, Osawa H, Satoh K, et al. Aberrant expression of CDX2 in Barrett's epithelium and inflammatory esophageal mucosa. J Gastroenterol 2003;38:14–22.
van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 2005;435:959–963.
Lorenz O, Duluc I, Arcangelis AD, et al. Key role of the cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J Cell Biol 1997;139:1553–1565.
Park J, Schulz S, Waldman SA . Intestine-specific activity of the human guanylyl cyclase C promoter is regulated by Cdx2. Gastroenterology 2000;119:89–96.
Yamamoto H, Bai Y-Q, Yuasa Y . Homeodomain protein CDX2 regulates goblet-specific MUC2 gene expression. Biochem Biophys Res Commun 2003;300:813–818.
Mesquita P, Jonckheere N, Almeid R, et al. Human MUC2 mucin gene is transcriptionally regulated by cdx homeodomain proteins in gastrointestinal carcinoma cell lines. J Biol Chem 2003;278:51549–51556.
Alkhoury F, Malo MS, Mozumder M, et al. Differential regulation of intestinal alkaline phosphatase gene expression by Cdx1 and Cdx2. Am J Physiol 2005;289:G285–G289.
Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 2010;139:2113–2123.
Mendelson J, Song S, Li Y, et al. Dysfunctional transforming growth factor-β signaling with constitutively active notch signaling in Barrett's esophageal adenocarcinoma. Cancer 2011;117:3691–3702.
Acknowledgements
We greatly appreciate the excellent technical support provided by Mrs Keiko Masuzaki. This work was supported in part by a grant-in-aid for Scientific Research (20590724) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Overexpression of Cdx2 by stimulation with bile acids induces activation of ATOH1 via Hes1 suppression during Barrett's epithelium development. These findings suggest that a transcriptional network related to Notch signaling and Cdx2 expression has an important role in the development of esophageal intestinal metaplasia.
Rights and permissions
About this article
Cite this article
Tamagawa, Y., Ishimura, N., Uno, G. et al. Notch signaling pathway and Cdx2 expression in the development of Barrett's esophagus. Lab Invest 92, 896–909 (2012). https://doi.org/10.1038/labinvest.2012.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2012.56
Keywords
This article is cited by
-
NACK and INTEGRATOR act coordinately to activate Notch-mediated transcription in tumorigenesis
Cell Communication and Signaling (2021)
-
Genome-wide tracts of homozygosity and exome analyses reveal repetitive elements with Barrets esophagus/esophageal adenocarcinoma risk
BMC Bioinformatics (2019)
-
Origins of Metaplasia in Barrett’s Esophagus: Is this an Esophageal Stem or Progenitor Cell Disease?
Digestive Diseases and Sciences (2018)
-
High expression of Claudin-2 in esophageal carcinoma and precancerous lesions is significantly associated with the bile salt receptors VDR and TGR5
BMC Gastroenterology (2017)
-
Reflux esophagitis and its role in the pathogenesis of Barrett’s metaplasia
Journal of Gastroenterology (2017)