Abstract
CD24 is a small, highly glycosylated cell surface protein that is linked to the membrane through a glycosyl-phosphatidylinositol anchor. It is overexpressed in many human carcinomas and its expression is linked to bad prognosis. Lately, lack or low expression of CD24 was used to identify tumor stem cells resulting in conflicting data on the usefulness of this marker. In many immunohistochemical studies, the mAb SN3b was used but the epitope and specificity of this antibody have never been thoroughly investigated. In other studies based mainly on cytofluorographic analysis, the mAb ML-5 was applied. In this study, we compared the epitope of mAb SN3b to the CD24 mAbs SWA-11 and ML-5 that both bind to the core protein of CD24. Using tissue microarrays and affinity-purified CD24 glycoforms, we observed only a partial overlap of SN3b and SWA11 reactivity. The mAb SN3b recognizes sialic acid most likely on O-linked glycans that can occur independently of the CD24 protein backbone. The SN3b epitope was not related to common sialylated cancer-associated glycan structures. Both SN3b epitope positive or negative CD24 glycoforms supported the binding of P-selectin and Siglec-5. In breast cancer, the SN3b reactivity was associated with bad prognosis, whereas SWA11 was not. In renal cell cancer, the SN3b epitope was completely absent but SWA11 reactivity was a prognostic factor. Our results shed new light on the tumorbiological role of CD24 and resolve discrepancies in the literature related to the use of different CD24 mAbs.
Similar content being viewed by others
Main
CD24 is a membrane glycoprotein with unusual lipid-like features.1 These characteristics are due to a small protein core of 27 (mouse) or 30 amino acids (human), extensive glycosylation, and the linkage to the cell membrane through a glycosyl-phosphatidylinositol (GPI) anchor.2, 3 In humans, CD24 is expressed by subpopulations of hematopoietic cells, regenerating muscle cells, keratinocytes and diverse tumor types such as breast, ovarian, nonsmall lung and pancreatic carcinomas.1 CD24 in combination with CD44 is frequently used as a marker for cancer stem cells. Initially discovered as a lymphoid differentiation marker, CD24 is also implicated as a crucial factor in certain forms of autoimmune disease.4 In multiple sclerosis or systemic lupus erythromatosis, CD24 was identified as a genetic modifier for risk and progression.4, 5 In this context, two CD24 polymorphisms were discovered that affect the expression levels of CD24 at the cell surface.4, 5
Changes in glycosylation have been extensively associated with cancer, and specific carbohydrate structures were used as markers for diagnosis and prognosis in different types of cancers. Usually, increased N-glycan branching is observed, whereas mucin-type O-glycans get shorter core 1-based (Galβ3GalNAc-Thr/Ser) (reviewed in Burchell et al6). Sialylated epitopes, such as sialyl-Lewisx (sLex NeuAcα2,3Galβ4(Fucα3)GlcNAc) or sialyl-Lewisa (sLea NeuAcα2,3Galβ3(Fucα4)GlcNAc) are increased and are relevant in cancer metastasis, such as colon and breast cancer (reviewed in Kannagi7). Other sialylated epitopes including sialyl-Tn (STn; NeuAcα2,6GalNAc-Ser/Thr), sialyl-3T (S-3T; NeuAcα2,3Galβ3GalNAc-Ser/Thr), sialyl-6T (S-6T; Galβ3(NeuAcα2,6)GalNAc-Ser/Thr) and disialyl-T (NeuAcα2,3Galβ3(NeuAcα2,6)GalNAc-Ser/Thr) are increased in O-glycans from mucins in several cancers, including breast cancer (reviewed in Burchell et al6 and Brockhausen8). Furthermore, there is a poor prognosis for breast cancer patients with STn. STn and MUC1 mucin, its most extensively studied carrier in cancer cells, have served as target for cancer immunotherapy in preclinical and clinical studies.9
CD24 is highly glycosylated and contains three potential N-glycosylation sites and several potential O-glycosylation sites. Recently, there were two reports in which the detailed structures of N- and O-glycans from mouse brain CD24 have been reported.10, 11 Brain type structures have been identified, which included α2,3-linked N-acetylneuraminic acid (NeuAc), Lewisx H antigens, bisecting N-acetylglucosamine in the N-glycans. On the other hand, the O-glycans consisted of mucin-type species (eg, S-3T antigen) or the O-mannosyl species, and contained α2,3-linked NeuAc, disialyl motifs, Lewisx sLex or HNK-1 epitopes. Carbohydrate structures of CD24 from tumor cells have not been described in detail in the literature; however, sLex from adenocarcinoma cells was found to mediate the interaction with P-selectin, therefore, underlying their rolling in vitro and in vivo.12, 13
In many carcinomas, CD24 expression has been linked to bad prognosis1 but the molecular basis for this is presently unknown. Importantly, not the expression at the plasma membrane, but rather the localization in cytoplasmic compartments showed the highest correlation with bad prognosis.1 In nearly all immunohistochemical studies, the mAb SN3b was used as it showed excellent abilities to detect CD24 on paraffin-embedded tumor tissues. However, the unique specificity of SN3b for CD24 was never properly investigated. Similar to many anti-carbohydrate mAbs, the SN3b mAb is an IgM and the antigenic determinants defined by this mAb include sialic acid residue(s) attached to the cells though a protein backbone.14 Blocking studies indicated that mAb SN3b indeed recognized CD24,14 but it was not excluded that the mAb might crossreact with other cell surface glycoproteins or might not detect all glycoforms of CD24.
In this study, we have examined the specificity of mAb SN3b for CD24 and compared it with the established CD24 mAbs ML-5 and SWA11. The latter mAb reacts with the protein core of CD24 and recognize a short leucine–alanine–proline sequence close to the GPI anchor.15, 16 We observed that mAb SN3b recognizes a carbohydrate epitope associated with some but not all forms of CD24 in carcinoma cells. The epitope was identified as an O-linked, sialic acid-containing moiety. In carcinoma cell lines, the reactivity of SN3b and SWA11 was not correlated and CD24 was expressed in the absence of SN3b reactivity. Conversely, in breast and prostate and renal carcinoma tissue sections, SN3b reactivity was observed independently of mAb SWA11. This pattern of binding suggested that the SN3b epitope can decorate other glycan moieties. Importantly, SN3b reactivity was correlated with poor prognosis in breast cancer, whereas SWA11 reactivity was not. These results shed new light on CD24 and a putative role of its associated glycans.
MATERIALS AND METHODS
Cells
Carcinoma cell lines were obtained from the Tumorbank of the German Cancer Research Center, Heidelberg, Germany. The B-lymphoma cell lines, Nalm-2 and Nalm-6, were obtained from Reinhard Schwartz-Albiez and Stefan Eichmüller, respectively (German Cancer Research Center, Heidelberg, Germany). The ovarian carcinoma cell line SKOV3ip was described before.17, 18 Cells were cultivated in DMEM supplemented with 10% FBS, penicillin/streptomycin, 10 mM glutamine at 37°C, 5% CO2 and 100% humidity.
Stable Expression of Human Fucosyltransferases In SKOV3ip Cells
Adherent SKOV3ip cell monolayers in six-well plates were transfected with pCDNA3.1/lacZ-V5 (Invitrogen) as control, and vectors coding different fucosyltransferases, pCDNA3.1/FUT3-V5,19 pCR3.1/FUT5, pCR3.1/FUT6 and pCR3.1/FUT7,20 using Lipofectamine Plus Reagent (Invitrogen), according to the manufacturer's protocol. Transfectants were selected with 2 mg/ml of Geneticin (G418 sulfate) (Invitrogen).
Chemicals and Antibodies
The mAb SWA11 and ML-5 to human CD24 were described.15, 21, 22 The mAbs SN3b and SN3 were obtained from Neolab (Thermo Fischer, Frankfurt, Germany). Mouse IgG anti-sialyl-Lea CA19-9 (192) was from Santa Cruz Biotechnologies. Mouse IgM anti-sialyl-Lex KM93 was from Chemicon (Millipore). Rabbit IgG anti-V523 was a kind gift of Professor Robert Doms, University of Pennsylvania. Goat anti-mouse IgM tetramethylrhodamine β-isothiocyanate (TRITC) conjugate was from Sigma. Goat anti-mouse IgG AlexaFluor594, goat anti-rabbit IgG AlexaFluor488 and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen. Recombinant Siglecs and selectins as Fc-fusion proteins were obtained from R&D (Wiesbaden, Germany).
Affinity Purification of CD24 and ELISA
This technique was described before.24, 25 Briefly, cell lysates were prepared in 2% Nonidet P-40 (NP40) and cleared by ultracentrifugation (30 min, 100 000 × g). Lysates were passed through a mouse IgG column to remove unspecific binding followed by a SWA11-coupled sepharose column. After extensive washing of the column, the bound antigen was elutated with 50 mM diethylamine/HCl (pH 11.5) containing 10 mM β-octylglycoside. Column fractions were neutralized and analyzed by ELISA using SWA11 mAb. Peak fractions were pooled and used for coating of ELISA plates at a 1:200 dilution in 50 mM carbonate buffer (pH 9.3). The procedure for ELISA with mAb or Fc-fusion proteins has been described.24 For biochemical analysis, an aliquot of the pooled fractions was precipitated with a fivefold volume of cold acetone for overnight at −20°C. The precipitated material was isolated by centrifugation (20 min at 5000 × g) and air dried.
Cytofluorographic Analysis
Cells were washed, resuspended in cold phosphate-buffered saline (PBS) containing 5% FBS and then incubated with mAb to CD24 for 30 min followed by washing and incubation for 20 min with PE-conjugated IgG secondary antibody (Dianova, Hamburg, Germany).26 For background binding controls, the primary mAb was omitted. Cells were analyzed with FACS Calibur (Becton Dickinson, Heidelberg, Germany). For data analysis, FlowJo software (Ashland, OR, USA) was used.
Immunofluorescence Microscopy
Cells grown on glass coverslips to approximately 80% confluency were washed with PBS containing 0.5 mM MgCl2, fixed with 4% (w/v) paraformaldehyde in PBS for 20 min and permeabilized with 0.1% (w/v) Triton X-100 for 15 min. Fixed cells were blocked with 1% of bovine serum albumin in PBS for 1 h, followed by incubations at room temperature of 2 and 1 h with primary and secondary antibodies, respectively. Antibodies were diluted in PBS containing 1% bovine serum albumin and washes were done with PBS. Primary antibody dilutions used were anti-sialyl-Lea at 1:2500, anti-sialyl-Lex at 1:160, mAb SN3b at 1:50 and anti-V5 at 1:500. Secondary antibody dilutions used were TRITC conjugate at 1:64, AlexaFluor594 and AlexaFluor488 conjugates at 1:500. Staining of the nucleus was carried out with the addition of DAPI at 1:2000 dilution. Coverslips were mounted in Airvol and examined with a Leica DMRB microscope (Leica Microsystems, Melbourne, VIC, Australia). Images were acquired using a COHU high-performance CCD camera coupled to the microscope and Leica QFISH software, with exposure times of 300 and 400 ms.
Glycosidase Hydrolysis
Hydrolysis of NeuAc from the purified CD24 was carried out by addition of 15 mU of neuraminidase from Vibrio cholerae or from Arthrobacter urefaciens (Roche) in 50 mM sodium acetate (pH 5.5) containing 4 mM CaCl2 and 50 mM sodium acetate (pH 50), respectively. For specific hydrolysis of α2,3-linked NeuAc 9 U of neuraminidase from Streptococcus pneumoniae (Prozyme, Glyko) were used following the supplier's instructions. For deglycosylation with peptide-N-glycosidase F (PNGase F) and endoglycosidase H (Endo H) (Roche), CD24 was boiled for 10 min in the presence of 0.2% (m/v) SDS and 1% (v/v) β-mercaptoethanol. After cooling down, 0.4% (w/v) NP40 was added followed by 1 U PNGase F in 50 mM sodium phosphate, 10 mM EDTA, pH 7.5, or 2.5 mU Endo H was added in 50 mM of sodium citrate pH 5.5. Complete protease inhibitors cocktail tablets 2% (m/v) (Roche) was added to all samples. The incubations were carried out at 37°C, overnight, or for 1 h for the neuraminidase from S. pneumoniae. Digestion with 15 mU endo-β-galactosidase from Bacteroides fragilis (QAbio), 1 mU O-glycosidase from Diplococcus pneumoniae (Roche), 4.32 mU β-galactosidase from bovine testes (Sigma) and 30 mU α-L-fucosidase from bovine kidney were all performed following the supplier's protocol.
Cell Lysis and Western Blot Analysis
Cell pellets were solubilized in lysis buffer (20 mM Tris/HCl (pH 8.0) containing 50 mM BOG or 1% Triton X-100, 10 mM NaF, 10 mM orthovanadate, 1 mM PMSF and 1 μg/ml of each leupeptin, aprotinin and pepstatin. Lysates were cleared by centrifugation and boiled with reducing or nonreducing SDS-sample buffer. Samples were separated on SDS-PAGE gels and transferred to Immobilon membranes using semi-dry blotting. After blocking with 5% skim milk in TBS or 3% bovine serum albumin in PBS (for SN3b), membranes were probed with primary antibodies followed by horseradish peroxidase-conjugated anti-mouse secondary antibody and ECL detection (GE Healthcare, Freiburg, Germany).
IHC
For comparative immunohistochemistry (IHC), three previously published tumor cohorts encompassing breast cancer,27 renal cell cancer (RCC) and prostate cancer28 were used. Automated IHC staining of formalin-fixed, paraffin-embedded (FFPE) tissue was carried out using Benchmark XT (Ventana, Tucson, AZ, USA). The mAb SWA-11 hybridoma supernatant against CD24 was diluted 1:5. DAB (Ventana) served as chromogen.
To detect CD24 with SN3b (final dilution 1:300), automated IHC staining of FFPE was carried out using the Bond™ automated IHC and in situ hybridization system (Leica Microsystems) using the Refine 30′/30′ detection system and the H2 10/95°C pretreatment protocol. To detect CD24 with SN3 (final dilution 1:100), the same detection system was used but heat-induced antigen retrieval was omitted. Afterward, the slides were briefly counterstained with hematoxylin and aqueously mounted.
Evaluation of IHC
For both mAbs the staining intensity was scored semiquantitatively, ranging from negative to strong (0, 1+, 2+, 3+). In addition, subcellular differentiation of staining patterns (membranous, cytoplasmic) was performed for SN3b as described in earlier studies.1 For SWA11, which invariably shows a cytoplasmic immunoreactivity in tumors, only total staining was considered.
Statistical Analysis
Statistics were calculated with SPSS V16. Spearman's rank correlation was used to compare immunoreactivity. Univariate survival statistics were calculated according to Kaplan–Meier with log-rank test.
RESULTS
Comparison of CD24 mAbs on Tumor Tissues
We initially compared the reactivity pattern by IHC of four mAbs to CD24 (SN3, SN3b, ML-5 and SWA-11) on a TMA containing 80 breast carcinoma tissue sections. The latter mAb was shown to react with the protein core of CD24.15 Although ML-5 and SWA11 showed a very high degree of similarity (correlation coefficient (CC) 0.776, P=0.001)), there was no correlation between SN3 or SN3b with SWA11 (SN3: CC=−0.024, P=0.84; SN3b: −0.114, P=0.315). In addition, there was no correlation between SN3 and SN3b detection (CC=0.131, P=0.282). These results prompted us to evaluate the widely used SN3b and SWA11 binding in more detail in other cohorts.
Breast cancer (n=183)
From our original SN3b-based study on CD24 expression in breast cancer (n=201), slides from 183 cases were available for further analysis and were immunostained with SWA11.27 For statistical comparison with SN3b, the original data set from 2002 was used; additional SN3b stainings were carried out for single cases.
In these 183 cases, 28 cases (15%) were completely negative for SN3b, 75 cases (41%) were weakly positive, 64 cases (35%) were moderately positive and 16 cases (9%) stained strongly. SN3b immunoreactivity showed no correlation with tumor grade (CC=0.001, P=0.988), pT stage (CC=0.138, P=0.062), ER status (CC=0.063, P=0.418) or HER2 overexpression (CC=0.059, P=0.466), but correlated to a positive pN status (CC=0.194, P=0.009).
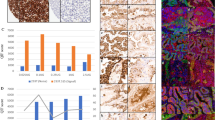
Using SWA11, 20 cases (11%) were negative, 38 cases (21%) were weakly positive, 52 cases (28%) were moderately positive and 73 cases (40%) stained strongly. SWA11 immunoreactivity did not correlate to pT stage (CC=0.113, P=0.127) but correlated with tumor grade (CC=0.224, P=0.002), nodal status (CC=0.149, P=0.044), ER status (CC=−0.159, P=0.040) and HER2 overexpression (CC=0.267, P=0.001). In breast cancer, SN3b and SWA11 intensity were not correlated (CC=0.092, P=0.216). Examples of congruently and noncongruently stained breast cancer cases are shown in Figure 1a.
Comparison of CD24 mAbs on carcinoma tissues. (a) Immunohistochemical staining of breast carcinoma sections using mAbs SN3b and SWA-11. Representative cases from a tissue microarray comprising 183 breast carcinomas are depicted to illustrate the different types of immunoreactivity obtained with both antibodies. The first panel (−/−) shows a case that is negative for both mAbs. The minor brownish shade seen for SWA11 does not justify to count this case as SWA11 positive, given the rather intense immunoreactivity else noted with this mAb. The second panel (−/+) shows completely SN3b-negative cases with strong SWA11 immunoreactivity. The third panel (+/−) shows the reverse: strong membranous and cytoplasmic SN3b staining in the absence of a significant SWA11 immunoreactivity. Lastly, in panel 4 a case strongly positive for both mAbs is shown. (b) Immunohistochemical staining of a renal carcinoma TMA (105 cases) with mAbs SN3b and SWA-11. Here, the overall weaker SN3b immunoreactivity is readily observable. Panel 1 (−/−) shows negative for both antibodies. Panel 2 (–/+) shows a moderate to strong SWA11 staining. Panel 3 (+/−) shows a weak SN3b staining, SWA11 is negative. Panel 4 (+/+) shows a weak SN3b staining, whereas SWA11 immunoreactivity is of moderate intensity. (c) Immunohistochemical staining of a prostate adenocarcinoma TMA (95 cases) with mAbs SN3b and SWA-11. In panel 1 (−/−) both cases are negative and in panel 2 (−/+) SN3b is negative, but SWA11 is weakly positive. Panel 3 (+/−) is weakly positive for SN3b but lacks SWA11 staining, and panel 4 (+/+) shows a moderate to strong staining with both mAbs. Magnification of all photographs is × 400.
RCC (n=105)
Using SN3b, 59 cases (56%) were completely negative, 39 (37%) were weakly positive, 5 (5%) were moderately positive and only 2 cases (2%) stained strongly. SN3b immunoreactivity showed no correlation with tumor grade (CC=0.150, P=0.127) or pT stage (CC=-0.082, P=0.406) but correlated weakly to a positive pM stage (CC=0.199, P=0.042).
Using SWA11, 21 cases (19%) were negative, 19 cases (18%) were weakly positive, 37 cases (35%) were moderately positive and 28 cases (27%) stained strongly. SWA11 immunoreactivity did not correlate to tumor grade (CC=0.127, P=0.195), pT- (CC=0.059, P=0.548) or pM stage (CC=0.148, P=0.132), but did correlate significantly and positively to SN3b staining (CC=0.262, P=0.007) (Figure 1b).
Prostate cancer (n=95)
Using SN3b, 46 cases (48%) were completely negative, 32 (34%) were weakly positive, 15 (16%) were moderately positive and only 2 cases (2%) stained strongly. SN3b immunoreactivity showed no correlation with Gleason's score (CC=0.063, P=0.549), preoperative PSA levels (CC=−0.039, P=0.720), pT stage (CC=−0.031, P=0.763) or margin status (R0/R1) (CC=0.112, P=0.284).
Using SWA11, 30 cases (32%) were negative, 33 (35%) were weakly positive, 25 (26%) were moderately positive and 7 cases (7%) stained strongly. SWA11 immunoreactivity showed no correlation with Gleason's score (CC=−0.059, P=0.572), preoperative PSA levels (CC=−0.184, P=0.090), pT stage (CC=−0.023, P=0.824) or margin status (R0/R1) (CC=0.100, P=0.339) (Figure 1c). The expression of both antibodies showed a trend toward a positive correlation (CC=0.197, P=0.056), but failed significance.
A statistical overview of overlapping and nonoverlapping cases for all three tumor entities under investigation is shown in Table 1. The rate of concordance is clearly tumor type dependent, ranging from 53% (RCC) to 78% (breast cancer).
The statistical analysis showed for all three tumor entities that both cytoplasmic and membranous SN3b staining was highly correlated to the total SN3b CD24 immunoreactivity of the initial analysis.
To extend these results by biochemical means, five randomly selected breast carcinoma tissues were solubilized in NP-40 and examined by western blot analysis using SN3b and SWA11 mAbs. Although all the tumor tissues showed strong reactivity with mAb SWA11, SN3b binding was only seen in part of the samples (Figure 2a) comigrating with the SWA-11 signal. It is interesting that mAb SN3b showed binding to protein bands that were not detected by SWA11 (see Figure 2a lane # 312). Collectively, the results suggested that SN3b reactivity can occur independently of SWA11 binding and vice versa and may also show a tumor entity-specific relationship.
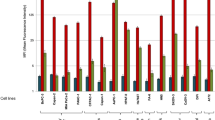
Comparison of CD24 mAbs on cancer tissues and cell lines. (a) Western blot analysis of breast carcinoma lysates. Tumor lysates (20 μg protein per lane) were analyzed with mAbs SN3b and SWA11 followed by peroxidase-conjugated goat anti-mouse IgG and IgM. < denotes a protein band only recognized by mAb SN3b. (b) FACS analysis of tumor cell lines. Cells were stained with the indicated mAbs (SWA-11, SN3b) to CD24 followed by PE-conjugated goat anti-mouse Ig secondary antibody (open curves). For background controls, the primary mAb was omitted (shaded curves). Note that SWA11 recognizes the protein core sequence of CD24. SN3b recognizes a carbohydrate epitope associated with CD24.
Comparison of CD24 mAbs on Cell Lines
When the distribution of SWA11 and SN3b epitopes was analyzed by cytofluorographic analysis on a panel of tumor cell lines, the SWA11 epitope was abundantly detected. Strong SN3b binding was only present in the B-lymphoma cell lines Nalm-2 and Nalm-6 (Figure 2b).
To examine whether both mAbs were indeed recognizing the same molecule, we carried out depletion experiments using Nalm-2 and Nalm-6 cells. Cell lysate were prepared and used for immunoprecipitation with SWA11-coupled sepharose. After removal of the sepharose-bound CD24, the lysates were again examined by western blot using mAbs SWA11 and SN3b. The depletion with SWA11 led to a clear reduction of SN3b reactivity (Figure 3a).
The SN3b epitope can decorate the CD24 antigen. (a) Depletion of cell lysates (Nalm-2 or Nalm-6) with mAb SWA11-coupled sepharose codepletes SN3b reactivity. Please note that the lanes showing SWA11 precipitates are heavily overloaded because of bound antigen. (b) Affinity-purified CD24 from the indicated cell lines was analyzed by silver staining and western blot analysis as indicated above. (c) Purified CD24 from SKOV3ip or Nalm-2 cells was used for ELISA with mAbs SWA11 and SN3b followed by peroxidase-conjugated goat anti-mouse IgG and IgM. (d) Purified CD24 from SKOV3ip or Nalm-2 cells was used for ELISA with recombinant Siglec or selectin–Fc fusion proteins complexed with peroxidase-conjugated goat anti-human IgG.
Analysis of Affinity-Purified CD24
To corroborate that the SN3b epitope was presented on the CD24 antigen, we performed affinity purification on SWA11 sepharose. By silver staining, the purified CD24 revealed multiple bands in the range of 30–80 kDa (Figure 3b) consistent with the high degree of N- and O-linked glycosylation.22 Western blot analysis revealed strong binding of SWA11 and SN3b to the purified CD24 from Nalm-2 cells (representing B-lymphoblastic cells) (Figure 3b). In contrast, CD24 from SKOV3ip cells (carcinoma cells) reacted only with SWA11 (Figure 3b). Similar results were obtained by ELISA (Figure 3c). These data suggested that mAb SN3b binds to an epitope that can associate with CD24 but is not expressed by all forms of CD24.
CD24 Supports Binding by P-Selectin and Siglec-5
We reported previously that CD24 can serve as a ligand for P-selectin.13 A recent study has shown that mouse CD24 can bind to Siglec-10,29 a member of the Siglec family of glycan-binding proteins that are differentially expressed on immune cells.30, 31 We investigated whether recombinant human Siglec-3, -5 and -10 could bind to purified human CD24 in ELISA. Only Siglec-5 showed strong binding to CD24 (Figure 3d). There was also strong binding of recombinant P-selectin but not E-selectin (Figure 3d) in agreement with our previous study.13, 24 Siglec-5 and P-selectin could not discriminate between the two glycoforms from SKOV3ip or Nalm-2 cells under ELISA conditions (Figure 3d).
Characterization of the SN3b Epitope
As CD24 is highly glycosylated, we argued that the SN3b epitope represented a post-translational modification of the protein core. Indeed, previous analysis had shown that the SN3b epitope included sialic acid,14 which indicated the requirement of glycans. We used affinity-purified CD24 from Nalm-2 cells to further characterize the glycosylation of the SN3b epitope. We observed that digestion with V. cholerae or A. ureafaciens neuraminidases that remove α2–3 and α2–6-linked NeuAc, abolished the SN3b epitope contrary to the SWA11 epitope that was left intact (Figure 4a). Digestion with neuraminidase from S. pneumoniae that is specific for α2–3-linked NeuAc abolished only part of SN3b binding (Figure 4a), which indicated that part of the NeuAc from the SN3b was α2,3 linked.
The SN3b epitope is sialic acid dependent and is present in O-glycans. (a) Purified CD24 from Nalm-2 cells was digested with neuraminidases from A. urefaciens (Au), V. cholerae (Vc) and S. pneumoniae (Sp), PNGase F and Endo H. *Indicates the control. CD24 was analyzed by western blot using mAbs SN3b and SWA-11. (b) SKOV3ip cells were stably transfected with vectors encoding FUT3 (SKOV3/FUT3), FUT6 (SKOV3/FUT6) and mock transfected (SKOV3/mock) as indicated, and the expression of sLea, sLex and SN3b epitopes was analyzed by immunofluorescence microscopy. Antibodies used were anti-sLea CA19-9 (192) (red), anti-V5 (green), anti-sLex KM93 (red) and anti-SN3b (SN3b) (red). Staining of the nucleus was carried out with DAPI (blue).
CD24 was digested with PNGase F, which removes N-glycans of the complex- and high-mannose type, and showed a downward shift of ∼13 kDa in western blot, which indicated that the N-glycosylation sites were occupied. However, there was still SN3b binding to the de-N-glycosylated form, suggesting that the epitope was most likely present in the O-glycans (Figure 4a). It can also be considered that the epitope could be present in the glycan core of the GPI anchor. Digestion with Endo H that removes high-mannose glycans did not cause a change in migration, showing that CD24 did not contain this type of structures (Figure 4a).
We investigated whether the sLea or sLex epitopes, which are increased in glycans from breast cancer6, 8 would be structurally related to the SN3b epitope. For this we overexpressed fucosyltransferases that participate in the biosynthesis of sialyl-Lewis determinants in the ovarian carcinoma cell line SKOV3ip, which has endogenously low or undetectable levels of these determinants.32 Stable overexpression of FUT3 produced high levels of sLea, whereas FUT6, followed by FUT7 and FUT5 produced high levels of sLex as detected by immunofluorescence microscopy (Figure 4b and data not shown). However, the SN3b antibody did not bind to these cell lines, which indicated that the SN3b epitope did not consist of sLea or sLex (Figure 4b).
The specificity of SN3b toward glycans was thoroughly investigated using the mammalian glycan array version 3.2 from the Consortium for Functional Glycomics (http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml). SN3b did not bind to any of the 406 glycans from the array (Supplementary Figure 1). The absence of binding to sLea (present in glycans 216 and 217) and to sLex (present in glycans 228–232, 373) corroborated the results presented in Figure 4b. Furthermore, SN3b was found not to bind STn (240 and 241), S-3T (222, 314), S-6T (320) and sialyl-dimeric Lex (231) that are also increased in cancer, which indicated that SN3b must be distinct from any of those sialylated O-linked glycans. In addition, CD24 was not sensitive to endo-β-galactosidase from B. fragilis or to β-galactosidase from bovine testes, which indicated, respectively, that SN3b was not present on poly-N-acetyllactosamine repeats and that it did not contain terminal β1,3/4-galactose. Furthermore, the signal was not abolished after digestion with α-L-fucosidase from bovine kidney nor with O-glycosidase, which indicated that the epitope did not contain fucose nor asialylated O-linked ±Galβ3GalNAc motifs (data not shown). Therefore, the results suggested that SN3b requires sialylated glycans either from the O-glycans or from the core of the GPI anchor.
Comparison of Prognostic Significance of CD24 mAb
Given the different staining pattern of SN3b and SWA11 on breast, renal cell and prostate carcinoma, we examined its association with clinical outcome. For survival analysis, all CD24 data sets were dichotomized in CD24-negative and -positive cases, as has been carried out in our previous analysis.33 All tumor cohorts have been previously published.28 In breast cancer, we observed as before in the minimally larger original cohort that SN3b positivity had a significant prognostic value (P=0.041). For SWA11 positivity, no prognostic value could be demonstrated (P=0.898) (Figure 5a).
SN3b and SWA-11 binding and prognostic significance. Staining data for SN3b and SWA-11 reactivity obtained from the (a) breast carcinoma cohort (183 cases), (b) renal cell carcinoma TMA (105 cases) and (c) prostate carcinoma TMA (95 cases) were correlated to clinical data of patient outcome (breast and renal cell cancer: overall survival, prostate cancer: PSA relapse-free survival), dotted line—CD24 negative, bold line—CD24 positive.
For RCC, no prognostic value of either antibody was found (Figure 5b). However, when both CD24 data sets were dichotomized according to the median (SN3b, -2: 0; SWA11: 2), SN3b did still not show any prognostic value (P=0.613), whereas high rates of SWA11 were significantly associated with shorter overall survival times (P=0.018). For prostate cancer, we could not detect any prognostic value of either antibody in this study (Figure 5c).
DISCUSSION
CD24 has gained attention as a prognostic marker in solid tumors and, in combination with CD44, is often used to define tumor stem cells.34 In the present report, we have compared the epitope specificity of CD24 mAbs. We find that (i) the frequently used mAb SN3b recognizes a glycan epitope that can occur independently of the CD24 protein core; (ii) the epitope of this mAb comprises sialic acid on O-linked glycans or at the glycan core of the GPI anchor; (iii) The SN3b epitope was not related to common sialylated cancer-associated glycan structures. Our findings have broad implications for diagnostic procedures in which mAbs to CD24 are used.
Our rationale to start analyzing CD24 in solid tumors came from the transcript-profiling study of prostate cancer, which identified CD24 mRNA as upregulated in approximately (change fold tumor or normal >2) 19% of cases.35 The functional data available at that time clearly hinted at a possible tumorbiological relevance of CD24. Its identification as a P-selectin ligand by Sammar et al12, 22, 24, 25 provided a possible mechanism for an increased metastatic potential of CD24-positive tumor cells. In addition, Senner et al36 had shown in an animal model of glioma that CD24-positive tumors grew significantly more aggressively. Until 2001, no paraffin-suitable CD24 antibodies were commercially available, which precluded an analysis of paraffinized archive tissues. This changed immediately with the launch of the paraffin-suitable monoclonal IgM CD24 antibody SN3b. This technically robust antibody easily allowed establishing IHC protocols for FFPE. The availability of a well-characterized cohort of ovarian cancer cases and the synchronous publication of CD24 as an upregulated transcript in ovarian cancer37 prompted us to analyze this tumor entity first. The strong and multivariately independent prognostic value found for cytoplasmic CD24 in this small tumor cohort, conferring a relative risk for disease progression of 7.7,38 was astounding but could in due course be reproduced, although at lower levels, for breast and prostate cancer, nonsmall cell lung cancer and colon cancer.27, 33, 39, 40
Since this first description of CD24 as a prognostic marker in a solid tumor in 2002,38 numerous studies have immunohistochemically analyzed CD24 expression in human tumors. A minimum of 27 studies, excluding our own, can be identified in PubMed (‘CD24 cancer immunohistochemistry’), encompassing all major tumor entities including esophageal,41 gastric42 and colon cancer,43 bladder44 and renal cancer,45 cholangiocarcinoma46 and endometrial carcinoma47 to name a few. In these studies, SN3b was by far (n=20, 74%) the most commonly used antibody to detect CD24. Other, rarely used CD24 antibodies were SN3 (n=2), SWA11 (n=1) and ML-5 (n=1). Three studies did not specify details concerning its CD24 antibody. The popularity of the CD24 antibody SN3b and the notion of possible discrepancies with other CD24 antibodies prompted us to conduct a systematic analysis of its specificity and its epitope. These are crucial aspects that have so far been neglected by the community of novel CD24 researchers coming from other fields such as cancer stem cell biology.
The perception of CD24 as a predominantly oncogenic gene, which is clearly advocated by the aforementioned studies has been blurred by the description of tumor stem cells in breast cancer by Al-Hajj et al34, which were defined by their negativity or low levels of CD24. These were apparently conflicting findings: a minority of tumorigenic and therefore assumedly aggressiveness conferring cells were according to Al-Hajj et al characterized by a loss of CD24 (the mAb to CD24 was not specified in this study but was most likely ML-5). In contrast, we and others48 found that tumors without CD24-positive cells had a considerably better prognosis (using mAb SN3b). There are several possible explanations for this discrepancy. First, the findings of Al-Hajj et al could be a mouse model artefact that does not translate into human biology. This interpretation was supported by the study of Abraham et al49, (using SN3b) which has failed to demonstrate that tumors bearing higher numbers of assumedly aggressive stem cell type (CD44+ and CD24–) cells have a more aggressive course. A serious methodological difference is that FACS usually examines the cell surface and thus is restricted to analyzing membranous CD24, whereas in IHC studies the cytoplasmic but not the membranous CD24 was of prognostic significance. Another, probably even more important issue that has been neglected so far is the specificity of the CD24 antibodies used in molecular and in situ studies and the comparability of their results. Cell line-based studies usually used CD24 antibodies that worked well in FACS systems but which are difficult to establish for IHC of FFPE materials. Possibly, both methods have investigated noncomparable epitopes leading to conflicting results.
This study provides substantial in vivo and in vitro evidence, that mAb SN3b does not identify the CD24 protein core itself, but binds to a glycan structure that decorates the CD24 molecule. However, this glycan is not present on all forms of CD24 and, possibly more important, can be present on other yet unidentified molecules irrespective of CD24. This explains the differences seen in the statistical evaluation of CD24 expression in the three tumor cohorts evaluated in this comparative study by the ‘classical’ SN3b and SWA11. SWA11 is a mouse monoclonal IgG antibody that specifically targets the CD24 protein core and that performs excellently on FFPE tissues, but which is not commercially available. The rates of CD24 positivity with SWA11 exceed those obtained with SN3b in all three tumor cohorts, which is consistent with the notion that SN3b epitope does not label all CD24 molecules. In addition, the often dichotomous membranous vs cytoplasmic staining seen with SN3b is uncommon with SWA11, which has a more uniform cytoplasmic pattern. It is interesting that the correlation analysis showed tumor type-dependent associations between the immunoreactivity of both antibodies, ranging from a fairly close correlation in RCC to a complete lack of significance in breast cancer. This is also mirrored in the survival analyses with SWA11 that differed from SN3b. Although for the three tumor entities SN3b immunoreactivity has been published as a prognostic marker,45, 50 we could not reproduce this for renal cell and prostate cancer, which might be sample size related. Conversely, in breast and prostate cancer, SWA11 immunoreactivity was not prognostic at all, whereas in RCC a prognostic value was seen. In an attempt to demonstrate specificity of the SN3b antibody, we reported before that a CD24-transfected cell line showed a higher degree of immunoreactivity than nontransfected cells by cytofluorographic analysis, which clearly supported detection of CD24 by SN3b and which was state of the art at that time.40 However, we now know that not all CD24 molecules are necessarily detected with the same sensitivity, probably dependent on glycosylation pattern, which is illustrated by the discrepancies we now noted by comparing an antibody directed against the protein core with the widely used SN3b, that is likely to detect a carbohydrate on CD24 that can also be expressed independently of CD24. It should be noted that a similar noncongruent staining of CD24 mAbs was noted previously on activated T lymphocytes.16
The SN3b-recognized epitope may be an important glycan structure in its own right. The results showed that SN3b requires sialylated glycans from CD24 for binding. The lack of binding to the most common sialylated epitopes from O-glycans that have been associated with different types of cancer, such as STn, S-3T, S-6T, sialyl-dimeric Lewisx sLea and sLex, suggested that the epitope is very unique and may constitute a highly specific and novel marker. It can also be admitted that because of the abundance and proximity of potential O-glycosylation sites of CD24 the epitope may include more than one O-glycan in a conformation that may also be affected by the peptide moiety of CD24. Finally, it may also be possible that the epitope is present in the glycan core of CD24, though less likely, as to our knowledge sialic acid has not been found in the core of several GPI anchors from human origin, even if its presence was detected in the core of the GPI anchors dipeptidase from pig kidney and PrPsc from hamster (reviewed by Paulick and Bertozzi51). Further structural study is needed to closer define the SN3b epitope at the molecular level.
During this study, we were also able to identify a novel CD24 ligand. To date, the only known ligand for CD24 is P-selectin expressed on activated platelets and endothelial cells22 and this interaction was shown to support the rolling of breast carcinoma cells.12 In this study, we show that purified CD24 also supports binding of recombinant Siglec-5. Siglecs are a group of sialic acid-binding proteins that are preferentially expressed on immune cells and have an essential role in immune regulation.30 Interestingly, recently it was reported that mouse CD24 could associate with high mobility group box 1 (HMGB1), heat shock protein 70 and heat shock protein 90 to prevent NF-κB activation through interaction with Siglec-10.29 It was shown that the Siglec-CD24 pathway was required to protect the host against a lethal response to pathological cell death.29 Our results show that human CD24 interacts with Siglec-5 rather than Siglec-10. Siglec-5 is expressed by macrophages, neutrophils and dendritic cells. It remains to be investigated whether the binding shown here by ELISA between purified proteins is of functional importance in the context of cells.
From our findings, we draw two major conclusions. First, it would be valuable to re-produce all CD24 studies originally conducted with the SN3b with the truly CD24-specific antibody SWA11 or ML-5 to validate the prognostic value of CD24. Second, the whole CD24 story impressively underscores the necessity to validate antibody specificity thoroughly before conducting larger expression studies using commercially obtained antibodies, which are often only insufficiently characterized.
References
Kristiansen G, Sammar M, Altevogt P . Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol 2004;35:255–262.
Kay R, Rosten PM, Humphries RK . CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J Immunol 1991;147:1412–1416.
Wenger RH, Ayane M, Bose R, et al. The genes for a mouse hematopoietic differentiation marker called the heat-stable antigen. Eur J Immunol 1991;21:1039–1046.
Wang L, Lin S, Rammohan KW, et al. A dinucleotide deletion in CD24 confers protection against autoimmune diseases. PLoS Genet 2007;3:e49.
Zhou Q, Rammohan K, Lin S, et al. CD24 is a genetic modifier for risk and progression of multiple sclerosis. Proc Natl Acad Sci USA 2003;100:15041–15046.
Burchell JM, Mungul A, Taylor-Papadimitriou J . O-linked glycosylation in the mammary gland: changes that occur during malignancy. J Mammary Gland Biol Neoplasia 2001;6:355–364.
Kannagi R . Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression—the Warburg effect revisited. Glycoconj J 2004;20:353–364.
Brockhausen I . Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep 2006;7:599–604.
Pinho S, Marcos NT, Ferreira B, et al. Biological significance of cancer-associated sialyl-Tn antigen: modulation of malignant phenotype in gastric carcinoma cells. Cancer Lett 2007;249:157–170.
Bleckmann C, Geyer H, Lieberoth A, et al. O-glycosylation pattern of CD24 from mouse brain. Biol Chem 2009;390:627–645.
Bleckmann C, Geyer H, Reinhold V, et al. Glycomic analysis of N-linked carbohydrate epitopes from CD24 of mouse brain. J Proteome Res 2009;8:567–582.
Aigner S, Ramos CL, Hafezi MA, et al. CD24 mediates rolling of breast carcinoma cells on P-selectin. Faseb J 1998;12:1241–1251.
Friederichs J, Zeller Y, Hafezi-Moghadam A, et al. The CD24/P-selectin binding pathway initiates lung arrest of human A125 adenocarcinoma cells. Cancer Res 2000;60:6714–6722.
Fukukawa T, Matsuzaki H, Haruta Y, et al. New monoclonal antibodies SN3, SN3a, and SN3b directed to sialic acid of glycoprotein on human non-T leukemia cells. Exp Hematol 1986;14:850–855.
Weber E, Lehmann HP, Beck-Sickinger AG, et al. Antibodies to the protein core of the small cell lung cancer workshop antigen cluster-w4 and to the leucocyte workshop antigen CD24 recognize the same short protein sequence leucine-alanine-proline. Clin Exp Immunol 1993;93:279–285.
Salamone MC, Rosselot C, Salamone GV, et al. Antibodies recognizing CD24 LAP epitope on human T cells enhance CD28 and IL-2 T cell proliferation. J Leukoc Biol 2001;69:215–223.
Runz S, Keller S, Rupp C, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol 2007;107:563–571.
Runz S, Mierke CT, Joumaa S, et al. CD24 induces localization of beta1 integrin to lipid raft domains. Biochem Biophys Res Commun 2008;365:35–41.
Sousa VL, Brito C, Costa J . Deletion of the cytoplasmic domain of human alpha3/4 fucosyltransferase III causes the shift of the enzyme to early Golgi compartments. Biochim Biophys Acta 2004;1675:95–104.
Grabenhorst E, Nimtz M, Costa J, et al. In vivo specificity of human alpha1,3/4-fucosyltransferases III-VII in the biosynthesis of LewisX and Sialyl LewisX motifs on complex-type N-glycans. Coexpression studies from bhk-21 cells together with human beta-trace protein. J Biol Chem 1998;273:30985–30994.
Jackson D, Waibel R, Weber E, et al. CD24, a signal-transducing molecule expressed on human B cells, is a major surface antigen on small cell lung carcinomas. Cancer Res 1992;52:5264–5270.
Aigner S, Sthoeger ZM, Fogel M, et al. CD24, a mucin-type glycoprotein, is a ligand for P-selectin on human tumor cells. Blood 1997;89:3385–3395.
Crystal AS, Morais VA, Pierson TC, et al. Membrane topology of gamma-secretase component PEN-2. J Biol Chem 2003;278: 20117–20123.
Sammar M, Aigner S, Hubbe M, et al. Heat-stable antigen (CD24) as ligand for mouse P-selectin. Int Immunol 1994;6:1027–1036.
Aigner S, Ruppert M, Hubbe M, et al. Heat stable antigen (mouse CD24) supports myeloid cell binding to endothelial and platelet P-selectin. Int Immunol 1995;7:1557–1565.
Schabath H, Runz S, Joumaa S, et al. CD24 affects CXCR4 function in pre-B lymphocytes and breast carcinoma cells. J Cell Sci 2006;119: 314–325.
Kristiansen G, Winzer KJ, Mayordomo E, et al. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res 2003;9:4906–4913.
Weber A, Kristiansen I, Johannsen M, et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer 2008;8:369.
Chen GY, Tang J, Zheng P, et al. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 2009;323:1722–1725.
Crocker PR, Paulson JC, Varki A . Siglecs and their roles in the immune system. Nat Rev Immunol 2007;7:255–266.
Varki NM, Varki A . Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest 2007;87:851–857.
Escrevente C, Machado E, Brito C, et al. Different expression levels of alpha3/4 fucosyltransferases and Lewis determinants in ovarian carcinoma tissues and cell lines. Int J Oncol 2006;29:557–566.
Kristiansen G, Pilarsky C, Pervan J, et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate 2004;58:183–192.
Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003;100:3983–3988.
Kristiansen G, Pilarsky C, Wissmann C, et al. Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J Pathol 2005;205:359–376.
Senner V, Sturm A, Baur I, et al. CD24 promotes invasion of glioma cells in vivo. J Neuropathol Exp Neurol 1999;58:795–802.
Welsh JB, Zarrinkar PP, Sapinoso LM, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001;98:1176–1181.
Kristiansen G, Denkert C, Schluns K, et al. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol 2002;161:1215–1221.
Kristiansen G, Schluns K, Yongwei Y, et al. CD24 is an independent prognostic marker of survival in nonsmall cell lung cancer patients. Br J Cancer 2003;88:231–236.
Weichert W, Denkert C, Burkhardt M, et al. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res 2005;11:6574–6581.
Sano A, Kato H, Sakurai S, et al. CD24 expression is a novel prognostic factor in esophageal squamous cell carcinoma. Ann Surg Oncol 2009;16:506–514.
Chou YY, Jeng YM, Lee TT, et al. Cytoplasmic CD24 expression is a novel prognostic factor in diffuse-type gastric adenocarcinoma. Ann Surg Oncol 2007;14:2748–2758.
Sagiv E, Memeo L, Karin A, et al. CD24 is a new oncogene, early at the multistep process of colorectal cancer carcinogenesis. Gastroenterology 2006;131:630–639.
Smith SC, Oxford G, Wu Z, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res 2006;66:1917–1922.
Lee HJ, Kim DI, Kwak C, et al. Expression of CD24 in clear cell renal cell carcinoma and its prognostic significance. Urology 2008;72:603–607.
Su MC, Hsu C, Kao HL, et al. CD24 expression is a prognostic factor in intrahepatic cholangiocarcinoma. Cancer Lett 2006;235:34–39.
Kim KH, Choi JS, Kim JM, et al. Enhanced CD24 expression in endometrial carcinoma and its expression pattern in normal and hyperplastic endometrium. Histol Histopathol 2009;24:309–316.
Mylona E, Giannopoulou I, Fasomytakis E, et al. The clinicopathologic and prognostic significance of CD44+/CD24(−/low) and CD44−/CD24+ tumor cells in invasive breast carcinomas. Hum Pathol 2008;39:1096–1102.
Abraham BK, Fritz P, McClellan M, et al. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res 2005;11:1154–1159.
Surowiak P, Materna V, Paluchowski P, et al. CD24 expression is specific for tamoxifen-resistant ductal breast cancer cases. Anticancer Res 2006;26:629–634.
Paulick MG, Bertozzi CR . The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry 2008;47:6991–7000.
Acknowledgements
This project was supported by a grant from the DKFZ-Bayer Schering Pharma Alliance to PA and GM. We express our gratitude to Dr Thomas Schlange, Bayer Schering, Pharma AG for continuous support, and David F Smith and Jamie Heimburg-Molinaro from the Core H unit of the Consortium for Functional Glycomics for SN3b glycan binding analysis. We thank Dr Harald Conradt, GlycoThera, Germany, for a gift of glycosidases and advice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kristiansen, G., Machado, E., Bretz, N. et al. Molecular and clinical dissection of CD24 antibody specificity by a comprehensive comparative analysis. Lab Invest 90, 1102–1116 (2010). https://doi.org/10.1038/labinvest.2010.70
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2010.70
Keywords
This article is cited by
-
Targeting CD24/Siglec-10 signal pathway for cancer immunotherapy: recent advances and future directions
Cancer Immunology, Immunotherapy (2024)
-
Expression of CD24 and Siglec-10 in first trimester placenta: implications for immune tolerance at the fetal–maternal interface
Histochemistry and Cell Biology (2017)
-
CD24 enrichment protects while its loss increases susceptibility of juvenile chondrocytes towards inflammation
Arthritis Research & Therapy (2016)
-
Membranous CD24 expression as detected by the monoclonal antibody SWA11 is a prognostic marker in non-small cell lung cancer patients
BMC Clinical Pathology (2015)
-
Aberrant immunostaining pattern of the CD24 glycoprotein in clinical samples and experimental models of pediatric medulloblastomas
Journal of Neuro-Oncology (2015)