Abstract
A key focus of research on pancreatic ductal adenocarcinoma (PDAC) is identifying new techniques to tailor gemcitabine and 5-fluorouracil treatments. Availability of tumor tissue is critical for the accurate assessment of gene expression, and laser microdissection (LMD) and primary cell cultures may be useful tools to separate tumor cells from the stromal reaction. The aim of this study was (1) to address the genetic profile relevant to drug activity and (2) to evaluate differences between microdissected and non-microdissected tumors, normal tissues, and primary cell cultures. Quantitative PCR of seven key genes was performed on mRNA from 113 microdissected and 28 non-microdissected tumors, a pool of normal tissues and four established primary cell lines. Protein expression was evaluated by western blot and immunocytochemistry and cytotoxicity by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. LMD allowed the analysis of 110 samples and revealed significant differences in mRNA levels between microdissected tumors and normal tissues, as well as between non-microdissected and microdissected tumors from the same patients. In contrast, primary cell lines showed similar expression profiles with respect to their respective microdissected tumors. In particular, expression levels of human equilibrative nucleoside transporter-1 and thymydilate synthase were significantly related to gemcitabine and 5-fluorouracil cytotoxicity. We conclude that LMD is a reliable technique for mRNA extraction, and allows detection of significant differences in the expression of specific target genes when compared to non-microdissected specimens and normal tissues. Moreover, expression levels in microdissected tumors are similar to those observed in primary tumor cell cultures, both at mRNA and protein level, and are related to drug chemosensitivity. The use of these ex vivo techniques for molecular analysis of tumors therefore appears to be of some value in implementing the clinical management of PDAC.
Similar content being viewed by others
Main
Pancreatic ductal adenocarcinoma (PDAC) is the fifth most common cause of death by cancer in the Western world, with an estimated incidence of more than 30 000 cases per year in the United States.1 Because of the anatomic location of the pancreas and the delay of clinical symptoms, in 85% of patients PDAC is detected at advanced stages, characterized by infiltration of proximal lymph nodes and vascular structures, as well as metastasis to the liver or peritoneum. A recent study reports a promising light scattering method for minimally invasive and early detection of pancreatic cancer.2 However, because of the extreme aggressive biological behavior and the lack of effective therapies, pancreas cancer has a poor prognosis even following surgical resection, and the 5-year survival for all stages of the disease remains <4%.3
Chemotherapy and radiation may only allow for a marginal increase in survival.3 Use of the deoxynucleoside analogue gemcitabine (2′,2′-difluorodeoxycytidine) has been compared favorably with the standard radiochemotherapy treatment for pancreatic cancer.4, 5 This molecule was able to inhibit the in vitro and in vivo growth of human pancreatic cells resistant to several other anticancer drugs, such as platinum compounds or 5-fluorouracil (5-FU), which are also widely used in the adjuvant and in the palliative second-line therapy of pancreatic cancer.6 However, high variability in clinical response has been observed and future directions to improve systemic therapy of PDAC should include the identification of molecular markers predictive of response, which may help in the selection of the best drugs for each patient.
Gemcitabine activity can be regulated by several proteins involved in drug transport, metabolism and mechanism of action, such as human equilibrative nucleoside transporter-1 (hENT1), deoxycytidine kinase (dCK), 5′-nucleotidase (5′-NT), cytidine deaminase (CDA), and ribonucleotide reductase subunit M1 (RRM1).7 Cells lacking hENT1 are highly resistant to gemcitabine,8 and pancreas cancer patients with the highest hENT1 protein or mRNA levels in tumor tissues have significantly longer overall survival (OS) after gemcitabine chemotherapy than patients affected by tumors with lower hENT1 expression.9, 10 Moreover, in the RTOG 9704 prospective randomized trial, hENT1 protein expression was associated with a statistically significant improvement in OS and disease-free survival in pancreatic cancer patients receiving gemcitabine.11 Similar genetic and molecular considerations might be relevant to the chemotherapeutic activity of 5-FU, and several studies investigated the association between clinical response and intratumoral gene expression of the key enzymes thymidylate synthase (TS)12, 13 and folylpolyglutamate synthetase (FPGS).14
PDAC usually incites an intense desmoplastic reaction,15, 16, 17 and recent studies have shown that the inflammatory and fibroblast cells in the stromal tissue exhibit different gene expression levels with respect to tumor cells.18, 19 Hence, laser microdissection (LMD) may be critical in enabling molecular analysis of PDAC and its biological characteristics.
In this study, we established a reliable and flexible technique for LMD in PDAC frozen sections, which, in combination with quantitative PCR, allowed a very fast, technically easy, and precise isolation of specific cell populations for mRNA analysis of key genes involved in drug transport and metabolism. This technique was used to compare mRNA expression pattern of these genes in micro- and non-microdissected tumor samples as well as in normal tissues. Furthermore, we found that the expression levels in microdissected tumors were similar to those observed in primary cell cultures, both at mRNA and protein level, and were related to the chemosensitivity of these cell lines to gemcitabine and 5-FU. These findings suggest the use of these ex vivo models for molecular analysis to implement the clinical management of PDAC.
MATERIALS AND METHODS
Patients Characteristics and Tissue Sampling
From December 2001 to July 2006, a total of 113 consecutive patients affected by PDAC were enrolled. Median age was 65 years (range, 22–83); 57 males and 56 females (Table 1). Surgical procedures consisted of pancreaticoduodenectomy (46.0%), distal (18.6%) or total pancreatectomy (7.1%), or biopsy (28.3%). Exocrine pancreatic tumors were histologically classified as proposed by the WHO (World Health Organization).20 As control, we also collected five cases of normal pancreatic tissues, obtained from the ‘Organ Donor Program’ at the University Hospital of Pisa (Italy). All tissue specimens analyzed in this study were obtained according to the institutional review board-approved procedures for consent from patients who underwent pancreatic resection at the Regional Referral Center for Pancreatic Disease Treatment, University Hospital of Pisa.
Immediately after surgical removal, tissue samples were frozen and stored in liquid nitrogen until LMD.
LMD
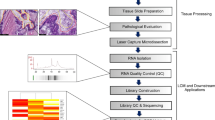
Frozen tissue sections (5 μm) were thawed, fixed in 75% ethanol, stained with haematoxylin/eosin, and dehydratated in 100% ethanol and xylene. Neoplastic cells were then dissected using the Leica LMD6000 instrument (Leica, Wetzlar, Germany), as described previously.21 Laser-captured cells were harvested in lysis buffer for RNA extraction. LMD was also used to obtain normal cells of epithelium ducts from five tissues pooled in a single cap, whereas in 28 cases RNA extraction was obtained from the whole tumor, without microdissection.
Isolation and Cultivation of Primary Cell Cultures
A total of 79 primary tumors were selected to set up primary cell cultures. The tumor tissue was washed extensively with phosphate-buffered saline and rinsed into approximately 1-mm3 cubes; these were plated onto 25 cm2 plastic flasks for primary tissue culture (PRIMARIA™ Tissue Culture Flask, Becton Dickinson, NJ, USA) containing a solution (1 mg/ml) of type XI Collagenase (SIGMA-ALDRICH, St Louis, MO, USA), in RPMI-1640 medium (Invitrogen Corporation, NY, USA), supplemented with 10% fetal calf serum, L-glutamine (2 mM), streptomycin (100 μg/ml) and penicillin (100 U/ml) at 37°C, in a humidified atmosphere containing 10% CO2. After 18 h, cells were harvested and seeded into a new flask. The medium was replaced every 3 days, until cell colonies were identified. All primary cell cultures were maintained by serial passages, once per week.
Cytotoxicity Studies
The cell growth inhibitory effect of gemcitabine and 5-FU was studied using the MTT assay both in primary cell cultures and in the established cell lines MIA PaCa-2, PANC-1, and Capan-1, cultured as described previously.10 For this purpose cells were plated at 104 cells per well in 96-well plates (Costar, Corning, NY, USA). After 24 h cells were treated with 5-FU (0.1–1000 μM) or gemcitabine (0.1–1000 nM) for 72 h. At the end of the incubation, cells were incubated for 3 h at 37°C in 50 μl MTT (final concentration, 0.5 mg/ml). Formazan crystals were dissolved in 150 μl of dimethylsulfoxide and the optical density was measured at 540 nm. Growth inhibition was expressed as the percentage of control (vehicle-treated cells) absorbance, corrected for absorbance before drug addition. The 50% inhibitory concentration of cell growth (IC50) was calculated by nonlinear least square curve fitting (GraphPad Prism, Intuitive Software for Science, San Diego, CA, USA).
Western Blot
To evaluate the protein expression of dCK, RRM1, and TS, total lysates were prepared from cells by lysis in buffer containing 50 mM Tris (pH 7.6), 20% (v/v) glycerol, 5 mM DTT, 0.5% (v/v) NP-40, and 4.0% (v/v) of a protease inhibitor cocktail. Lysates were sonicated and centrifuged. The protein-containing supernatant was collected and protein content was determined using a Bradford assay. In each lane of a minigel system (Bio-Rad, Hertfordshire, UK) 20 μg of proteins were loaded and separated on a 10% SDS–PAGE gel, followed by blotting onto a nitrocellulose membrane. The membrane was preincubated in blocking buffer for 1 h, whereas the mouse anti-human monoclonal TS antibody (1:100, TS-106 antibody, NeoMarkers, Fremont, CA, USA) and polyclonal mouse anti-human dCK antibody (1:500; Abnova, Taipei, Taiwan) were added overnight, at 4°C. After washing in TBS-T, the blots were incubated for 1 h with specific horseradish peroxidase-labelled secondary antibodies (1:2000; DakoCytomation, Glostrup, Denmark). Antibody binding was detected using enhanced chemiluminescence (Amersham Pharmacia Biotech, Milan, Italy) and measured by densitometric scanning software (Kontron, Eching, Germany). As a loading control expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined using a specific antibody (1:10 000, Chemicon International, Temecula, CA, USA).
Immunocytochemistry
The cells were grown in Chamber Slides System (Lab-Tek, IL, USA) in a humidified incubator at 37°C with 5% CO2. After 48 h the cells were fixed with acetic acid and methanol solution (ratio 1:3) at room temperature for 10 min. Immunocytochemistry (ICC) was performed using a specific monoclonal mouse anti-human RRM1 antibody (Chemicon, Hampshire, UK; overnight incubation and 1:30 dilution). The cells were then stained with avidin-biotin-peroxidase complex (Ultramarque™ HRP Detection kit, Greenwood, AR, USA). Negative controls were obtained replacing the primary antibody with buffer. The sections were reviewed and scored blindly by comparing the staining of tumor cells vs adjacent fibroblasts derived from pancreas tumors by two pathologists (DC and LEP). As there are no validated scoring systems to interpret immunocytochemical staining for RRM1, we used a system based on staining intensity and on the number of stained cells. With regard to the intensity score, if the intensity was comparable to the adjacent fibroblasts, intensity was classified as 0; if the cells stained weakly, intensity was classified as 1+; if the cells stained moderately, intensity was classified as 2+; and if the cells stained strongly, intensity was classified as 3+. With regard to the number of cells, if none of the cells was stained the score was 0, if less than 25% of the cells were stained the score was 1+, if 25–50% of the cells was stained the score was +2, and if more than 50% of the cells were stained, the score was 3+.
RNA Extraction
RNA was extracted from normal pancreatic ducts, primary and established cell cultures, microdissected and non-microdissected tumor tissue using the QiaAmp RNA mini-Kit (Qiagen, San Diego, CA, USA), and dissolved in 10 mM dithiothreitol in RNase-free water. RNA yields and integrity were checked by optical density at 260 nm with a UV-Vis Kontron Uvikon 940 spectrophotometer (Kontron, Milan, Italy), whereas testing for contamination by protein or by organic compounds, thiocyanates and phenolate ions was performed by measuring absorbance at 280 and 230 nm, respectively.
Quantitative PCR Analysis
After determining RNA quality by reverse transcription–PCR (RT–PCR) amplification of the widely used housekeeping gene GAPDH, from 50 to 500 ng RNA was reverse transcribed and the resulting cDNA was amplified by the 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA), as described previously.10
Forward and reverse primers and probes were designed using Primer Express 2.0 (Applied Biosystems) based on dCK (NM_000788), 5′-NT (NM_012229), CDA (NM_001785), TS (NM_001071), and FPGS (NM_001018078) gene sequence obtained from the Genbank, whereas primers and probes for RRM1 (NM_001033) and hENT1 (NM_004955) were obtained from Applied Biosystems Assay-on-Demand products (Hs00168784 and Hs00191940).
To determine the primer concentrations that give the best reproducibility (detected as minimum standard deviation (s.d.) between threshold cycle (CT) values assessed in quadruplicate experiments), we prepared four PCR reactions with all the combinations of forward and reverse primers at 300 and 900 nM concentrations and demonstrated that the optimal primer concentration was 300 nM for both forward and reverse primers for dCK, 5′-NT, and TS and 900 nM for CDA and FPGS. A validation experiment was also carried out to demonstrate that efficiencies of the target and reference (GAPDH) were approximately equal, using a relative standard curve method with several dilutions of the cDNA obtained from Q-PCR Human Reference Total RNA (Stratagene, La Jolla, CA, USA). As all calculations were based on the CT of the amplification plot in triplicate experiments, and as the higher the expression, the lower is the CT, data are expressed as GAPDH/target gene ratio, in order to give a direct comparison of gene expression level. Specimens were amplified in triplicate with appropriate non-template controls, and the coefficient of variation was <1% for all replicates.
Statistical Analysis
All experiments were performed in triplicate and were repeated at least three times. Data were expressed as mean values±s.d. and were analyzed by Student's t-test, using the GraphPad Prism software (GraphPad Prism, Intuitive Software for Science). The Pearson's/Spearman's correlation test and regression analysis were used to demonstrate the relationship between gene and protein expression profile and chemosensitivity; the level of significance was P<0.05.
RESULTS
Cell Cultures
From 79 primary tumors selected to set up primary cell cultures, we obtained 8 (12.7%) primary cell cultures (LPc006, LPc028, LPc033, LPc053, LPc067, LPc103, LPc111, and LPc167). RNA extracted from these primary cell cultures was used for gene expression analysis. In particular, in this study we used only the most stable primary cell cultures, after the 20th passage (LPc006, LPc028, LPc067, and LPc167 cells).
LMD and RNA Extraction
LMD was performed on 113 PDAC samples including 81 cases analyzed in a previous pharmacogenetic study,10 and four tumor specimens from which the primary cell cultures described above were derived. LMD was also performed on five specimens from normal ducts, and the cells from these tissues were used to obtain a normal tissue pool. For each sample, LMD allowed to pick up specimens of 5000 cells, which were harvested directly into the cap of a 200 μl microfuge tube. The precision of the narrow focus of the laser beam resulted in the capture of individual cells with high degree of accuracy and extremely low risk of contamination (Figure 1). In 28 randomly selected PDAC cases, the sampling was also performed on whole tissue, without LMD, using 3 cryostatic sections (thickness of 40 μm).
RNA was extracted from all these tumor samples, as well as from the normal tissue pool and from a pool of 106 cells of the four primary cell cultures described above, as well as from MIA PaCa-2, PANC-1, and Capan-1 cell lines. The analysis of protein contamination by absorbance reading demonstrated that most samples were pure, showing an optimal 260/280 ratio (1.8–2.0).
mRNA Gene Expression
After determining RNA quality using RT–PCR amplification of the housekeeping gene GAPDH, a total of 110 microdissected samples were evaluable, whereas three subjects had no suitable RNA for gene expression analysis. In these samples, quantitative assessment of the results of the gene expression analysis showed that mRNA expression of CDA was detectable in all the 110 samples, whereas the mRNA expression of dCK, 5′-NT, RRM1, hENT1, TS, and FPGS was detectable in 109, 106, 104, 109, 96, and 72 specimens, respectively.
The plot in Figure 2 shows the variability of the gene expression observed across the cohort of all the pancreatic cancer patients subjected to transcription analysis. Quantitative PCR data showed that the variability of triplicate CT values in individual samples was always lower than 0.3% of mean values, and allowed to detect a moderate inter-patient variability, with mean values±s.d. ranging from 0.953±0.069 (5′-NT) to 0.841±0.162 (FPGS).
It is worth noting that the gene expression profile differed significantly between WHO grade 1/2 (n, 48) and grade 3 (n, 33) samples. Indeed, hENT1 mRNA levels in WHO grade 3 specimens were significantly lower with respect to those measured in grade 3 PDAC samples (1.254±0.208 vs 1.151±0.215, respectively, P=0.035; Table 2). However, histological grading was not associated with significant differences in dCK, 5′-NT, CDA, RRM1, TS, and FPGS mRNA levels (Table 2).
The mRNA expression of dCK, 5′-NT, CDA, RRM1, hENT1, TS, and FPGS was also detected in the microdissected duct samples, and the statistical analysis revealed significant differences in mRNA expression levels between microdissected tumor tissues and normal pancreatic tissues for all the studied determinants (Table 3).
Similarly, the comparison of dCK, 5′-NT, CDA, RRM1, hENT1, TS, and FPGS expression levels among the non-microdissected tumor tissues and the respective microdissected samples with the paired Student's t-test demonstrated a meaningful statistical variation in 6 out of the 7 genes analyzed (Figure 3). In particular, a significant difference in mRNA expression values between microdissected and non-microdissected specimens was observed for dCK (1.000±0.112 vs 0.880±0.055; number of pairs, 27; P<0.001), CDA (0.940±0.083 vs 0.895±0.117; number of pairs, 28; p, 0.047), RRM1 (0.932±0.060 vs 0.891±0.040; number of pairs, 22; P=0.003), hENT1 (1.183±0.261 vs 0.834±0.112; number of pairs, 26; P<0.001), TS (1.025±0.116 vs 0.895±0.079; number of pairs, 27; P<0.001), and FPGS (0.903±0.064 vs 0.851±0.052; number of pairs, 23; P=0.003), whereas no statistically significant variation was detected for 5′-NT (0.919±0.062 vs 0.913±0.083; number of pairs, 26; P>0.05).
Finally, the PCR analysis of dCK, 5′-NT, CDA, RRM1, hENT1, TS, and FPGS showed similar gene expression profiles in the four established primary cell cultures (Figure 4), with differences in mean gene expression values ranging between 0.9% (variation of RRM1 mRNA expression in the LPc006 cell line and its respective microdissected tumor) and 22.8% (variation of hENT1 mRNA expression in the LPc028 cell culture and respective microdissected tumor). However, in most cases differences of gene expression values observed in cell lines with respect to those quantified in their respective microdissected tumor tissues were less than 5%.
In Vitro Studies
A dose-dependent inhibition of cell growth was observed after both gemcitabine and 5-FU treatment, with IC50s of 7.2±1.3 nM and 176.2±64.0 μM (LPc006), and 1.3±0.2 nM and 107.3±20.2 μM (LPc028), 5.5±0.1 nM and 94.9±24.9 μM (LPc067), and 9.7±0.1 nM and 136.2±8.3 μM (LPc167), respectively (Figure 5a and b). Similar results were observed in the commercial pancreatic cancer cell lines, MIA PaCa-2, PANC-1, and Capan-1 cells, with IC50s of 5.9±0.8, 17.9±1.8, and 7.2±1.3 nM, and 153.3±23.9, 201.4±24.5, and 146.1±11.5 μM for gemcitabine and 5-FU, respectively. Therefore, among the primary cell lines, LPc067 and LPc028 were the most sensitive cell lines to gemcitabine and 5-FU, respectively, whereas LPc006 were the most resistant cells to 5-FU, and LPc167 were the least sensitive cells to gemcitabine.
Results of in vitro studies. Representative curves of growth inhibitory effects of 5-FU (a) and gemcitabine (b); 72-h exposure. Protein expression as determined by western blot and immunocytochemistry: one representative blot (c) out of at least three independent western blot is shown for TS, dCK, and GAPDH protein expression, whereas four representative slides of the RRM1 staining are shown for the primary cell cultures (d). Left upper and lower panel show the slides of positive control (fibroblasts) and negative control (epithelial cells without primary antibody), respectively. Original magnification of controls, × 40; original magnification of LPc006 and LPc167, × 20. Correlation of hENT1 mRNA levels with gemcitabine chemosensitivity (e). Comparison between protein and expression levels of dCK (f) and TS (g), and correlation between TS expression and 5-FU chemosensitivity (g).
TS, dCK, and RRM1 expression were also studied at the protein level, using western blotting and ICC. Representative examples are shown in Figure 5c. These analyses showed relevant differences in dCK and TS expression among the four PDAC primary cell cultures. In particular, TS protein expression was twofold lower in LPc067 than in LPc006 cells, whereas TS levels in LPc028 and LPc167 cells were comparable. Similarly, as shown in Figure 5c, the LPc006 cells showed the highest dCK protein levels, whereas the faintest bands were observed in the extracts of LPc028 and LPc167 cells, and the lowest dCK expression was detected in the LPc067 cells.
Upon immunohistochemical examination, neoplastic cells showed variable RRM1 staining, which was related to their gene expression. In comparison with the fibroblasts (Figure 5d, left upper panel), the LPc006 cells, characterized by the higher mRNA levels, presented strong (+3) and diffuse (+3) staining (Figure 5d, right upper panel), whereas cells with an intermediate level of RRM1 mRNA (LPc028) showed moderate positivity (intensity score, +2, number of cells score, +3). Finally, cells with a slightly lower RRM1 mRNA expression (LPc067 and LPc167) had an intensity score of +2, whereas the number of stained cells was +2 (Figure 5d, right lower panel).
The variability in the expression of genes involved in the action of both gemcitabine and 5-FU in the PDAC cells may be involved in the different sensitivity to drug treatment. Although the small sample size of cell lines used in this study precluded the assessment of the predictive value of gene expression data as validated determinants of drug sensitivity, a clear correlation was found between the IC50 values of gemcitabine and the mRNA expression of hENT1 (R2=0.931, P=0.035, Figure 5e), ie, the lower chemosensitivity of the LPc167 cells to gemcitabine might depend on the lower mRNA expression of hENT1, with respect to the other cell lines (Figure 4). Similarly, the LPc006 cell line showed the highest expression of TS, associated with the lowest activity of 5-FU, despite a high expression of FPGS. In contrast, the 5-FU-sensitive LPc067 cells showed the lowest TS expression. Indeed, a significant correlation was also found between TS mRNA expression and 5-FU cytotoxicity (R2=0.839, P=0.042). To further investigate the relationship between gene expression and drug sensitivity, we also evaluated hENT1 and TS mRNA levels in MIA PaCa-2, PANC-1, and Capan-1 cells. In agreement with the previous data, the highest and the lowest expression of hENT1 (ie, 1.05 and 0.68 arbitrary units (a.u.), calculated as mean relative mRNA expression with respect to GAPDH) were detected in the most and in the least sensitive cell lines (ie, MIA PaCa-2 and PANC-1 cells, respectively). Furthermore, the hENT1 mean expression values from all the seven studied cells significantly correlated with gemcitabine sensitivity (R2=0.803, P=0.037). The analysis of TS mRNA levels in MIA PaCa-2, PANC-1, and Capan-1 cells (1.08, 1.25, and 0.93 a.u., respectively) also confirmed the significant correlation between TS mRNA expression and 5-FU cytotoxicity (R2=0.908, P=0.010, in the seven studied cells).
However, no correlations were found between dCK, CDA, 5′-NT, and FPGS mRNA gene expression and drug sensitivity in this panel of seven PDAC cells.
TS and dCK protein were measured with western blot in primary cell cultures and the analysis of band density demonstrated a significant link between both dCK and TS mRNA and protein expression in control cells, with r2 values of 0.877 and 0.869, respectively (P<0.05). However, the dCK expression was not correlated to gemcitabine chemosensitivity (Figure 5f), whereas 5-FU sensitivity was correlated to both TS mRNA and protein levels (Figure 5g).
DISCUSSION
High-throughput technology is continually being invented and refined to allow increasingly sensitive molecular analysis of tissue samples. As new cutting-edge quantitative expression analytical procedures and instruments are developed, more rigorous demands are placed on sample preparation techniques. Using LMD in combination with these sensitive analytical methods, investigators may obtain more accurate molecular data and address previously unanswerable questions arising from samples characterized by different cellular populations.
This technology is crucial to avoid several problems in gene expression studies in samples from adenocarcinomas of the exocrine pancreas. Indeed, this malignancy is characterized by a dense desmoplastic stromal component, with islets of neoplastic cells representing only a fraction of the tumor mass.22 Contiguous non-neoplastic areas of the pancreas may be affected by dysplastic changes or show chronic pancreatitis depending on whether there is distal obstruction of the pancreatic duct.23 Furthermore, PDAC originates from pancreatic ductal epithelium, a cell type that accounts for less than 5% of pancreatic tissue. For these reasons, the use of contiguous normal human pancreas tissue from resection specimens may be inappropriate as control. Indeed, previous studies demonstrated that, in comparison with normal ductal pancreatic cells obtained through an organ donor program, PDAC tissue specimens revealed differentially expressed genes belonging predominantly to the stromal component of the tumor.24 Significant differences in gene expression were also found between normal pancreas and tumor-cell microdissected samples.18
Therefore, in order to accurately measure the expression levels of drug-related genes in tumor cells, homogenous populations of neoplastic ductal cells from freshly frozen samples of pancreatic cancer patients were obtained by the LMD technique. In order to compare mean gene expression values of gemcitabine and 5-FU determinants in tumor and normal tissues, we used normal ductal pancreatic cells obtained from healthy individuals. The analysis of differences in the gene expression in long-time stabilized primary cell culture with respect to their primitive tumors was aimed at evaluating the possible role of this preclinical model for new therapeutic approaches, including pharmacological modulation of expression of genes involved in drug activity.
In agreement with previous studies,23, 24, 25 the high precision and affordability of the LMD technology allowed us to obtain samples containing about 95% of tumor cells, and provided RNA samples of good quantity and quality. The integrity of extracted RNA is essential for gene expression analyses and it is crucial if used in studies involving the pancreas, an organ with high endogenous nuclease activity.26 Hence, we examined the quality of the RNA extracted from each specimen by both absorbance reading, showing an optimal 260/280 ratio, and preliminary PCR amplification of GAPDH.
The successive validated quantitative PCR analysis was a reliable, sensitive, and specific technique to assess mRNA gene expression, even when sample material was limited. This enabled the measurement of mRNA gene expression in 110 out of 113 microdissected samples. It is noteworthy that techniques used in the present study allowed the accurate analysis of tissue gene expression within 6 h after sample delivery to the laboratory.
The comparison of 28 microdissected and non-microdissected tumor tissues revealed significant differences in expression levels of genes potentially affecting gemcitabine and 5-FU activity. These results are in agreement with the data obtained in previous studies on differences between neoplastic tissue and normal tissue or between neoplastic tissue and chronic pancreatitis,19 and might lead to clinically relevant applications. Indeed, several in vitro and in vivo studies demonstrated the fundamental role of molecular determinants involved in gemcitabine uptake and metabolism, such as hENT1 and dCK,27 whereas recent pharmacogenetic studies revealed that TS and hENT1 are prognostic factors in 5-FU-treated13 and gemcitabine-treated PDAC patients.10 In particular, in our previous analysis, we detected a significant correlation between gene expression values of hENT1 and clinical outcome in 81 gemcitabine-treated patients, using microdissected tumor specimens.10 In contrast, no correlation was observed between OS and dCK mRNA expression in microdissected pancreatic cancer specimens. Similarly, both the mRNA and the protein levels of dCK were not related to clinical outcome in non-small-cell lung cancer (NSCLC) patients treated with gemcitabine-based chemotherapy.28, 29, 30
Further analysis of gene expression profile in PDAC tumors according to their histological grade demonstrated that WHO grade 3 neoplasm displayed significantly lower hENT1 mRNA levels than grade 1/2 tumors. In view of the relevant role of hENT1 in the response to gemcitabine, these results suggest that the sensitivity profile to gemcitabine may change with respect to histological grade of neoplasms. Several studies demonstrated that histopathological grading could be useful for predicting prognosis in PDAC patients.31, 32 The multivariate analysis performed in our previous study showed that both disease grading and hENT1 expression are significant independent prognostic factors of PDAC.10 However, a recent prospective study reported that hENT1 protein expression was associated with a statistically significant improvement in OS and disease-free survival in pancreatic cancer patients receiving gemcitabine, but not in those not receiving gemcitabine.11
In our study a statistically significant difference was also documented comparing the microdissected tumor tissue and the microdissected normal tissue, as well as between the microdissected tumor tissue and the non-microdissected tumor tissue. The expression levels of dCK, 5′-NT, and CDA were gradually reduced from normal ductal epithelium to microdissected tumor tissue and, finally, to non-microdissected tumor. Similar results were reported by a previous study performed with LMD and quantitative PCR to evaluate differences among PDAC, normal pancreatic tissue, and stroma.18 This analysis demonstrated that mean levels of expression of molecules involved in cellular trophism were lower in tumor with respect to normal tissues. However, we found higher mean levels of RRM1, hENT1, TS, and FPGS in microdissected tumors with respect to both non-microdissected tumors and normal tissues. The proteins studied in our samples belong to the normal protein pattern of human cells, and the elevated mRNA gene expression levels of several of these proteins in the normal pancreatic duct cells may be explained by the functional activity of these cells. Indeed, the normal epithelium may synthesize different types of proteins in comparison to tumor cells, which devote the greatest part of cellular metabolism to proliferation. In contrast, the lowest values of expression found in most non-microdissected samples might be justified by the major amount of stroma, constituted by a mix of fibroblasts, inflammatory cells and normal pancreatic parenchyma.
Previous studies showed no significant differences between TS expression in colorectal tumor and normal mucosa,33 whereas the present study showed a different profile of TS in tumor and normal pancreas, suggesting that the highest expression of TS may reduce 5-FU antitumor activity. In contrast, other data suggested that upregulation of hENT1 may occur in certain solid tumors,34 and the highest expression of hENT1 detected in PDAC samples with respect to normal pancreas specimens may improve gemcitabine uptake and its therapeutic effect, even if other studies did not support the present results.35
Finally, although the mean difference of expression levels between non-microdissected and microdissected tumors is above 20%, we found a mean difference of only 5% in gene expression values in the comparison between primary cell cultures and their respective microdissected tumors.
These results suggest that both the LMD and the establishment of primary cell culture, with different methodological approaches, succeeded in eliminating the stroma, which can mask the true expression of several genes in PDAC cells.
The LPc167 cells showed the lowest sensitivity to gemcitabine and this finding was associated with the lower mRNA expression of hENT1 than in the other cell lines. In contrast, the LPc067 cells, the most sensitive to gemcitabine, showed the highest hENT1 gene expression. These results are in agreement with previous studies showing that the basal expression levels of hENT1 were significantly correlated with the IC50 values for gemcitabine in 22 NSCLC cell lines or with pancreatic carcinoma and biliary tract carcinoma cell lines.36, 37 Other data suggest that the sensitivity of pancreatic cancer cells to gemcitabine is determined by the ratio of gene expression of several factors involved in gemcitabine transport and metabolism, including hENT1, dCK, and RRM1.38, 39 Similarly, the LPc006 cell line had the highest expression of TS, associated with the lowest activity of 5-FU, whereas the 5-FU-sensitive LPc067 cells had the lowest TS expression. Thus, the results of the present in vitro studies provide further evidence to sustain the importance of mRNA level measurement in target tissue for a tailored chemotherapy.
Furthermore, the results concerning the significant relationship between mRNA and protein expression of dCK, RRM1, and TS are in agreement with previously published data in different cell lines and xenografts.40, 41, 42 These findings further support the use of quantitative PCR, which is a highly sensitive technique and a valuable tool for tumor cell-specific analysis of gene expression, even when the sample material is limited.
Therefore, both LMD and PCR technologies, and the primary cell cultures may play a pivotal role in future pharmacogenetic studies on the gene expression of molecular determinants of drug activity. In particular, a major issue in the next years will be the validation of these biomarkers in several prospective trials. Recent trials of customized chemotherapy based on genetic markers have been carried out in NSCLC and promising pharmacogenetic markers are gaining momentum.43, 44 Hopefully, validated biomarkers to select PDAC patients most likely to respond to tailored chemotherapeutic protocols will be established in the near future.
In conclusion, this study demonstrates that (1) LMD is a reliable and non-destructive technique for RNA extraction, (2) the selection of tumor cells by LMD allows detection of significant differences between the expression of several genes potentially involved in drug activity with respect to expression levels observed in non-microdissected specimens, as well as in normal tissues, and (3) the expression levels of microdissected tumor samples are similar to those detected in primary cell cultures, both at mRNA and protein level. We also demonstrate that hENT1 and TS expression levels are related to chemosensitivity in vitro. Hence, the use of these ex vivo models for molecular tumor analysis may be of particular value in implementing the management of chemotherapy in patients affected by PDAC.
References
Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. Cancer J Clin 2007;57:43–66.
Liu Y, Brand RE, Turzhitsky V, et al. Optical markers in duodenal mucosa predict the presence of pancreatic cancer. Clin Cancer Res 2007;13:4392–4399.
Li D, Xie K, Wolff R, et al. Pancreatic cancer. Lancet 2004;363:1049–1057.
Abbruzzese JL . New applications of gemcitabine and future directions in the management of pancreatic cancer. Cancer 2002;95 (Suppl 4):941–945.
Burris III HA, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1995;15:2403–2413.
Saif MW . Pancreatic cancer: highlights from the 42nd annual meeting of the American Society of Clinical Oncology 2006. J Pancreas 2006;7:337–348.
Giovannetti E, Mey V, Nannizzi S, et al. Pharmacogenetics of anticancer drug sensitivity in pancreatic cancer. Mol Cancer Ther 2006;5:1387–1395.
Mackey JR, Mani RS, Selner M, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res 1998;58:4349–4357.
Spratlin J, Sangha R, Glubrecht D, et al. The absence of Human Equlibrative Nucleoside Transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res 2004;10:6956–6961.
Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of Human Equilibrative Nucleotide Transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res 2006;66:3928–3935.
Farrell JJ, Garcia M, Lai R, et al. Human ENT1 is predictive of response to gemcitabine treatment in patients with pancreatic cancer: Results from the RTOG 9704 Prospective Randomized Trial. Pancreas 2007;35:401–402.
Takamura M, Nio Y, Yamasawa K, et al. Implication of thymidylate synthase in the outcome of patients with invasive ductal carcinoma of the pancreas and efficacy of adjuvant chemotherapy using 5-fluorouracil or its derivatives. Anticancer Drugs 2002;13:75–85.
Hu YC, Komorowski RA, Graewin S, et al. Thymidylate synthase expression predicts the response to 5-fluorouracil-based adjuvant therapy in pancreatic cancer. Clin Cancer Res 2003;9:4165–4171.
Kidd EA, Yu J, Li X, et al. Variance in the expression of 5-fluorouracil pathway genes in colorectal cancer. Clin Cancer Res 2005;11:2612–2269.
Simpson-Haidaris PJ, Rybarczyk B . Tumors and fibrinogen. The role of fibrinogen as an extracellular matrix protein. Ann NY Acad Sci 2001;936:406–425.
Muerkoster S, Wegehenkel K, Arlt A, et al. Tumor stroma interactions induce chemoresistance in pancreatic ductal carcinoma cells involving increased secretion and paracrine effects of nitric oxide and interleukin-1beta. Cancer Res 2004;64:1331–1337.
Ryu B, Jones J, Hollingsworth MA, et al. Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res 2001;61:1833–1838.
Ketterer K, Rao S, Friess H, et al. Reverse trascription-PCR analysis of Laser-Captured cells points to potential paracrine and autocrine action of neurotrophins in pancreatic cancer. Clin Cancer Res 2003;9:5127–5136.
Guweidhi A, Kleef J, Giese N, et al. Enhanced expression of 14-3-3sigma in pancreatic cancer and its role in cell cycle regulation and apoptosis. Carcinogenesis 2004;25:1575–1585.
Kloeppel G, Longnecker DS, Capella C, et al. Histological Typing of Tumors of the Exocrine Pancreas: World Health Organization International Histological Classification of Tumors. Springer-Verlag: Berlin, 1996.
Fukuschima N, Sato N, Prasad N, et al. Characterization of gene expression in mucinos cystic neoplasms of the pancreas using oligonucleotide microarrays. Oncogene 2004;10:9042–9051.
Solcia E, Capella C, Kloeppel G . Tumors of the Pancreas. Atlas of Tumor Pathology, 3rd edn. Fascicle 20. AFIP: Washington DC, USA, 1997.
Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, et al. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 2002;29:4587–4594.
Nakamura T, Furukawa Y, Nakagawa H, et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene 2004;13:2385–2400.
Burgermeister R . New aspects of laser microdissection in research and routine. J Histochem Cytochem 2005;53:409–412.
Peracaula R, Cleary KR, Lorenzo J, et al. Human pancreatic ribonuclease 1: expression and distribution in pancreatic adenocarcinoma. Cancer 2000;89:1252–1258.
Kroep JR, Loves WJ, van der Wilt CL, et al. Pretreatment deoxycytidine kinase levels predict in vivo gemcitabine sensitivity. Mol Cancer Ther 2002;1:371–376.
Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res 2004;10:4215–4219.
Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res 2004;10:1318–1325.
Seve P, Mackey JR, Isaac S, et al. cN-II expression predicts survival in patients receiving gemcitabine for advanced non-small cell lung cancer. Lung Cancer 2005;49:363–370.
Yeo CJ, Cameron JL, Sohn TA, et al. Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg 1997;226:248–257.
Yamamoto S, Tomita Y, Hoshida Y, et al. Prognostic significance of activated Akt expression in pancreatic ductal adenocarcinoma. Clin Cancer Res 2004;10:2846–2850.
Amatori F, Di Paolo A, Del Tacca M, et al. Thymidylate synthase, dihydropyrimidine dehydrogenase and thymidine phosphorylase expression in colorectal cancer and normal mucosa in patients. Pharmacogenet Genomics 2006;16:809–816.
Goh LB, Mack P, Lee CW . Nitrobenzylthioinosine-binding protein overexpression in human breast, liver, stomach and colorectal tumor tissues. Anticancer Res 1995;15:2575–2579.
Pennycooke M, Chaudary N, Shuralyova I, et al. Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem Biophys Res Commun 2001;280:951–9959.
Achiwa H, Oguri T, Sato S, et al. Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small-cell lung cancer. Cancer Sci 2004;95:753–777.
Mori R, Ishikawa T, Ichikawa Y, et al. Human equilibrative nucleoside transporter 1 is associated with the chemosensitivity of gemcitabine in human pancreatic adenocarcinoma and biliary tract carcinoma cells. Oncol Rep 2007;17:1201–1205.
Nakano Y, Tanno S, Koizumi K, et al. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer 2007;96:457–463.
Giovannetti E, Mey V, Danesi R, et al. Synergistic cytotoxicity and pharmacogenetics of gemcitabine and pemetrexed combination in pancreatic cancer cell lines. Clin Cancer Res 2004;10:2936–2946.
Sigmond J, Loves WJ, Kroep JR, et al. Quantitative real time PCR of deoxycytidine kinase mRNA by Light Cycler PCR; in relation to enzyme activity. Nucleosides Nucleotides Nucleic Acids 2004;23:1347–1350.
Bergman AM, Eijk PP, Ruiz van Haperen VW, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res 2005;65:9510–9516.
Peters GJ, Backus HH, Freemantle S, et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta 2002;1587:194–205.
Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol 2006;24:4731–4737.
Rosell R, Cobo M, Isla D, et al. Pharmacogenomics and gemcitabine. Ann Oncol 2006;17 (Suppl 5):v13–v16.
Acknowledgements
DC was supported by research grants from the Ministero dell’Istruzione, Università e Ricerca (Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale, PRIN 2005). EG received the L’Orèal-UNESCO Award for Women in Science 2007 (V Italian edition) for a project entitled ‘Pharmacogenetics of pancreas cancer’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Funel, N., Giovannetti, E., Del Chiaro, M. et al. Laser microdissection and primary cell cultures improve pharmacogenetic analysis in pancreatic adenocarcinoma. Lab Invest 88, 773–784 (2008). https://doi.org/10.1038/labinvest.2008.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2008.40
Keywords
This article is cited by
-
A Critical Comparison of Rejection-Based Algorithms for Simulation of Large Biochemical Reaction Networks
Bulletin of Mathematical Biology (2019)
-
TGF-β induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression
Nature Communications (2018)