Abstract
Analysis of our microRNA (miRNA) expression signatures of human cancers based on RNA sequencing have shown that both strands of pre-miR-150, miR-150-5p (the guide strand) and miR-150-3p (the passenger strand), are significantly reduced in cancer tissues. We have investigated the functional significance of both strands of pre-miR-150 in cancer cells. The aim of this study was to investigate the antitumor function of these miRNAs and how these miRNAs regulated oncogenic targets in esophageal squamous cell carcinoma (ESCC). Ectopic expression studies demonstrated that both strands of pre-miR-150 miRNA inhibited ESCC cancer cell migration and invasion, indicating that both miR-150-5p and miR-150-3p acted as antitumor miRNAs. A combination of genome-wide gene expression analyses and in silico database searches showed that SPOCK1 (SPARC/osteonectin, cwcv and kazal-like domains proteoglycan 1) was a candidate target of miR-150-5p and miR-150-3p in ESCC cells. Luciferase reporter assays showed that SPOCK1 was directly regulated by these miRNAs. Silencing of SPOCK1 by small interfering RNA inhibited cancer cell migration and invasion. Overexpression of SPOCK1/SPOCK1 was confirmed by real-time PCR methods and immunohistochemistry. Taken together, downregulation of both strands of pre-miR-150 and overexpression of SPOCK1 are involved in ESCC pathogenesis. The involvement of passenger strand miRNAs in the regulation of cancer cell aggressiveness is a novel concept in RNA research.
Similar content being viewed by others
Introduction
Esophageal cancer is the eighth most common cause of cancer worldwide and the sixth most frequent cause of cancer death.1 In Japan, esophageal cancer is the thirteenth most common cause of cancer, with an estimated 22 800 new cases in 2016.2 Esophageal cancer is categorized into two subtypes based on histology: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EADC).3, 4 These two cancers greatly vary in the etiological factors and geographic distribution.3, 5, 6 Approximately 90% of esophageal cancers are diagnosed as ESCC and mainly occur in Central Asia, North and Central China and Japan.3 The main risk factors of ESCC are tobacco use, alcohol consumption and drinking hot beverages.5 Owing to ESCC’s aggressive nature, the overall 5-year survival rate is <25% due to diagnosis in later stages accompanied by local invasion and distant metastasis.3, 4
Understanding the initiation and progression of ESCC requires analysis of gene regulation. In that regard, microRNAs (miRNAs) are non-coding RNA molecules that act as fine tuners of gene expression, modulating more than 30% of protein-coding genes in a sequence-specific manner.7 Notably, a single miRNA can regulate a large number of RNA transcripts in human cells.8 Therefore, aberrantly expressed miRNAs can disrupt RNA networks, leading to cancer cell development, metastasis and drug resistance.9
We have identified a number of antitumor miRNAs, and these miRNAs regulated novel cancer pathways, including those in ESCC.10, 11 Past studies showed that several miRNAs were aberrantly expressed in ESCC cells and appeared to function as oncogenes or antitumor miRNAs.12 Our studies showed that miR-375 was significantly reduced in cancer tissues and ectopic expression of miR-375 inhibited cancer cell aggressiveness through targeting of MTDH and MMP13.11
Recently, analyses of miRNA expression signatures of human cancers by RNA sequencing revealed that several miRNA passenger strands and guide strands were significantly reduced in cancer tissues.13, 14, 15, 16 Based on the expression signatures, we have investigated the functional significance of passenger strands of miRNAs in cancer cells. Our data showed that miR-144-3p and miR-145-3p were significantly downregulated in cancer cells, and ectopic expression of these miRNAs inhibited cancer cell aggressiveness by targeting several oncogenes.17, 18, 19 Our data indicated that passenger strands of miRNAs, miR-144-3p and miR-145-3p, actually acted as antitumor miRNAs. The involvement of passenger strand miRNAs in the regulation of cancer cell aggressiveness is a novel concept in RNA research.
In this study, we have focused on the dual strands of pre-miR-150, miR-150-5p (guide strand) and miR-150-3p (passenger strand), because these miRNAs were annotated as downregulated miRNAs in our expression signature of head and neck squamous cell carcinoma (HNSCC), using hypopharyngeal SCC specimens.20 Hypopharynx and esophagus are neighboring organs and it is considered that gene expression signatures are similar. Our previous studies showed that two expression lists of downregulated miRNAs were extremely similar by signatures of HNSCC and ESCC.13, 16, 21 The functional significance of miR-150-3p and its target genes in cancer cells is largely unknown. The aim of this study was to investigate the antitumor function of these miRNAs and how these miRNAs regulated oncogenic targets in ESCC.
Materials and methods
Cancer and normal tissue samples from ESCC patients
Clinical specimens were collected from 20 ESCC patients, including 10 non-cancerous tissues. Patients at the Kagoshima University Hospital, Kagoshima, Japan from 2010 to 2014 underwent primary surgical treatment and were pathologically confirmed to harbor ESCC. Our study was approved by the Bioethics Committee of Kagoshima University; written prior informed consent and approval were obtained from all patients. The clinicopathological characteristics of the patients are shown in Table 1.
Cell culture and RNA extraction
We used two ESCC cell lines: TE-8, which was moderately differentiated, and TE-1, which was highly differentiated. Both were provided by the Riken BioResourse Center (Tsukuba, Ibaraki, Japan). Cells were maintained in RPMI with 10% fetal bovine serum and penicillin according to standard culture procedures.
Total RNA from clinical specimens and cell lines was isolated using ISOGEN (Nippon Gene, Toyama, Japan) according to the manufacturer’s protocol.
Quantitative real-time reverse transcription-PCR
The procedure for quantitative real-time reverse transcription-PCR quantification was described previously.17, 18, 19 The expression levels of miR-150-5p or miR-150-3p (assay ID: 000473, 002637; Applied Biosystems, Foster City, CA, USA) were analyzed by TaqMan qRT-PCR assays (TaqMan MicroRNA assays; Applied Biosystems), and RNU48 (assay ID: 001006) was used for normalization. TaqMan probes and primers for SPOCK1 (assay ID: Hs00270274_m1; Applied Biosystems) and GUSB (the internal control; assay ID: Hs00939627_ml; Applied Biosystems) were used for gene expression analysis.
Transfection with mature miRNAs and siRNAs
The following mature miRNAs were used: Ambion Pre-miR miRNA precursor for hsa-miR-150-5p and miR-150-3p (product ID: PM10070, PM12324; Applied Biosystems). The following small interfering RNAs (siRNAs) were used: Stealth Select RNAi siRNA, si-SPOCK1 (cat. nos HSS110150 and HSS110151; Invitrogen, Carlsbad, CA, USA) and negative control miRNA/siRNA (P/N: AM17111; Thermo Fisher Scientific, Waltham, MA, USA). RNAs were incubated with Opti-MEM (Invitrogen) and Lipofectamine RNAiMax transfection reagent (Invitrogen), as described previously.17, 18, 19
miRNA incorporate into RISC by Ago2 immunoprecipitation assay
TE-8 cells were transfected with 10 nM miRNA by reverse transfection and plated in 10-cm plates at 1 × 105 cells per ml. After 48 h, immunoprecipitation was performed using a microRNA Isolation Kit, Human Ago2 (Wako, Osaka, Japan) according to the manufacturer’s protocol. Expression levels of miRNAs bound to Ago2 were measured by TaqMan RT-qPCR. miRNA expression data were normalized to the expression of miR-26a (product ID: 000404; Applied Biosystems), which was not affected by miR-150-5p or miR-150-3p.
Cell proliferation, migration and invasion assays
TE-8 and TE-1 cells were transfected with 10 nM miRNAs or siRNAs by reverse transfection. Cell proliferation, migration and invasion assays were performed as described previously.17, 18, 19
Screening of miR-150-5p and miR-150-3p target genes using in silico analysis and gene expression data
To identify miR-150-5p and miR-150-3p target genes, a combination of genome-wide gene expression and in silico analyses was conducted as described previously.17, 18, 19 The microarray data were deposited in the GEO repository under accession number GSE93290. Next, we selected putative miRNA target genes using TargetScanHuman ver.7.1 (June, 2016 release, http://www.targetscan.org/vert_71/) databases and the GEO database (GSE20347). Our strategy for identification of miR-150-5p and miR-150-3p target genes is shown in Figure 2.
Western blot analysis
Anti-human SPOCK1 rabbit polyclonal immunoglobulin G (1:1000; HPA007540; Sigma-Aldrich, St Louis, MO, USA) was used as a primary antibody. Anti-human glyceraldehyde 3-phosphate dehydrogenase mouse monoclonal immunoglobulin G (1:5000; 010-25521; Wako Pure Chemical Industries, Osaka, Japan) was used as an internal loading control.
Plasmid construction and dual luciferase reporter assays
Partial wild-type sequences of the 3′-untranslated region (UTR) of SPOCK1 containing the miR-150-5p or the miR-150-3p target site (miR-150-5p: positions 182–188 of the SPOCK1 3′-UTR; miR-150-3p: positions 1477–1483, 1749–1756 and 2593–2599 of the SPOCK1 3′-UTR) or sequences with a deleted miR-150-5p or miR-150-3p target sites were inserted into the psiCHECK-2 vector (product ID: C8021; Promega, Madison, WI, USA) as described previously.17, 18, 19, 20
Immunohistochemistry
Tumor samples were fixed with 10% formaldehyde in phosphate-buffered saline, embedded in paraffin and sectioned into 4-μm-thick slices. The sections were incubated with rabbit polyclonal anti-SPOCK1 immunoglobulin G (1:200; HPA007540; Sigma-Aldrich) at 4 °C overnight. The procedure for immunohistochemistry was described previously.20, 22
Statistical analysis
Relationships between two or three variables and numerical values were analyzed using the Mann–Whitney U-test or Bonferroni-adjusted Mann–Whitney test. Spearman’s rank test was used to evaluate the correlations between the expression levels of miR-150-5p, miR-150-3p and SPOCK1. Expert StatView version 5.0 (SAS Institute, Cary, NC, USA) was used in these analyses.
Results
The expression levels of miR–150–5p and miR–15–3p in ESCC specimens and cell lines
We assessed the expression levels of miR-150-5p and miR-150-3p in ESCC tissues, normal epithelial tissues and ESCC cell lines (TE-8 and TE-1) by quantitative real-time reverse transcription-PCR. Expression levels of miR-150-5p and miR-150-3p were significantly downregulated in cancer tissues and cancer cell lines compared with normal tissues (P<0.05; Figure 1a). Spearman’s rank test showed a positive correlation between the expression levels of miR-150-5p and miR-150-3p (P=0.0002, R=0.634; Figure 1b).
Expression levels of miR-150-5p and miR-150-3p in ESCC clinical specimens and functional assays in miR-150-5p- and miR-150-3p-transfected ESCC cell lines. (a) Expression levels of miR-150-5p in ESCC clinical specimens and cell lines. RNU48 was used as an internal control. (b) Correlation between expression levels of miR-150-5p and miR-150-3p in clinical specimens (R=0.634, P=0.0002). (c) Cell proliferation was determined by XTT assay 72 h after transfection with miR-150-5p or miR-150-3p. *P<0.0001. (d) Cell migration activity was determined by migration assay. *,P<0.0001. (e) Cell invasion activity was determined by Matrigel invasion assay. *,P<0.0001.
To elucidate molecular mechanisms of low expression of miR-150-5p and miR-150-3p in ESCC cells. Expression levels of miR-150-5p in ESCC cells was significantly elevated by the demethylation agent, 5-aza-dc treatment. In contrast, expression of miR-150-3p was moderately elevated by 5-aza-dc treatment (data not shown). In addition, our recent study of HNSCC cells showed that expression levels of both strands of pre-miR-150 were recovered by 5-aza-dc treatment.20 These data suggested that DNA methylation might cause silencing of miR-150-5p and miR-150-3p in ESCC cells.
Effects of ectopic expression of miR–150–5p and miR–150–3p on cell proliferation, migration and invasion assays in ESCC cell lines
To evaluate the effect of miR-150-5p and miR-150-3p in ESCC, we transfected precursor miR-150-5p and miR-150-3p into ESCC cell lines (TE-8 and TE-1). Cell proliferation was significantly inhibited in miR-150-5p- or miR-150-3p-transfected TE-8, but not in TE-1 (P<0.0001; Figure 1c). Cell migration and invasion assays demonstrated that cell migration and invasion were significantly inhibited by miR-150-5p or miR-150-3p transfection in both TE-8 and TE-1 in comparison with mock or miR-control transfectants (P<0.0001; Figures 1d and e). These results showed that both miR-150-5p and miR-150-3p could have antitumor functions in ESCC.
Both miR-150-5p and miR-150-3p were incorporated into RISC in ESCC cells
We hypothesized that both miR-150-5p and miR-150-3p may be incorporated into and function as part of the RISC structure. To test this hypothesis, we performed immunoprecipitation with antibodies targeting Ago2, which has a central role in the RISC. After transfection with miR-150-5p or miR-150-3p, Ago2-bound miRNAs were isolated, and quantitative real-time reverse transcription-PCR was carried out to determine whether miR-150-5p and miR-150-3p bound to Ago2 (Figure 2a).
miR-150-5p and miR-150-3p were incorporated into RISC. (a) Schematic illustration of miRNA detection method. Isolation of RISC-incorporated miRNAs by Ago2 immunoprecipitation. (b) Expression levels of miR-150-5p and miR-150-3p after transfection with miR-150-3p following immunoprecipitation by Ago2 (*P<0.0001).
After transfection with miR-150-5p and immunoprecipitation by anti-Ago2 antibodies, miR-150-5p levels were significantly higher than those of mock- or miR-control-transfected cells and those of miR-150-3p-transfected TE-8 cells (P<0.0001; Figure 2b). Similarly, after transfection with miR-150-3p and immunoprecipitation by anti-Ago2 antibodies, miR-150-3p levels were significantly higher than those of mock- or miR-control-transfected cells and those of miR-150-5p-transfected TE-8 cells (P<0.0001; Figure 2b).
Identification of miR-150-5p and miR-150-3p coordinately regulated genes in ESCC cells
To better understand how the downregulation of miR-150-5p and miR-150-3p promoted tumor aggressiveness, we searched miR-150-5p and miR-150-3p target genes using gene expression analyses and in silico analyses. First, we performed gene expression analyses using a miR-150-5p- or a miR-150-3p-transfected ESCC cell line (TE-8). A total of 7988 genes were downregulated (log2 ratio<−0.5) in miR-150-5p-transfected TE-8, and 6728 genes were downregulated in miR-150-3p-transfected TE-8 in comparison with the control. Using TargetScanHuman ver.7.1, we found that 1596 genes had miR-150-5p binding sites and 768 genes had miR-150-3p binding sites. Of those, 153 genes had both miR-150-5p and miR-150-3p binding sites, and 12 genes were upregulated in ESCC, as determined using ESCC gene expression data (GEO accession number: GSE20347; Figure 3). SPOCK1 was the most downregulated gene in miR-150-3p transfectants. Thus, we proposed that SPOCK1 was a target gene of miR-150-5p and miR-150-3p in ESCC (Table 2).
The strategy for identification of miR-150-5p and miR-150-3p target genes. Gene expression analyses revealed that 7988 genes and 6728 genes were downregulated by miR-150-5p and miR-150-3p transfected into TE-8 cells, respectively. Next, we merged the data for putative target genes in the TargetScan database (release 7.1). A total of 153 genes were identified as both miR-150-5p and miR-150-3p target genes. Among them, we examined the upregulated genes in ESCC clinical specimens using a GEO database (GSE: 20347). Finally, a total of 12 genes were listed as candidates of both miR-150-5p and miR-150-3p regulation in ESCC cells.
We showed that several putative targets regulated by miR-150-5p and miR-150-3p in ESCC cells independently (Supplementary Tables 1 and 2). These data are effective to explain antitumor roles of these miRNAs in ESCC cells.
Direct regulation of SPOCK1 expression by miR-150-5p and miR-150-3p in ESCC cells
We carried out luciferase reporter assays with a vector that included the 3′-UTR of SPOCK1 to confirm that miR-150-5p and miR-150-3p directly regulated SPOCK1 in a sequence-dependent manner. The TargetScanHuman database predicted that there was one binding site for miR-150-5p in the 3′-UTR of SPOCK1 (positions 182–188); it also predicted the presence of three binding sites for miR-150-3p in the 3′-UTR of SPOCK1 (Figure 4a). Co-transfection with pre-miR-150-5p or pre-miR-150-3p significantly reduced luciferase activity in comparison with mock and miR-control (P<0.05; Figure 4a).
Direct regulation of SPOCK1 by miR-150-5p and miR-150-3p in ESCC cell lines. (a) Putative miR-150-5p or miR-150-3p binding sites in the 3′-UTR of SPOCK1 mRNA. Luciferase reporter assays using vectors encoding putative miR-150-5p or miR-150-3p target sites of the SPOCK1 3′-UTR (positions 182–188, 1477–1483, 1749–1756 and 2593–2599) for both wild-type and deletion-type constructs. Renilla luciferase values were normalized to firefly luciferase values. *P<0.0001. (b) Expression levels of SPOCK1 mRNA 72 h after transfection with miR-150-5p or miR-150-3p. (c) SPOCK1 protein expression 72 h after transfection with miR-150-5p or miR-150-3p.
To confirm that miR-150-5p and miR-150-3p regulated SPOCK1, we evaluated expression levels of SPOCK1 mRNA and protein using quantitative real-time reverse transcription-PCR and western blotting. Expression levels of SPOCK1 were downregulated in miR-150-5p or miR-150-3p transfectants (P<0.0001 or P<0.05; Figures 4b and c).
Effects of silencing SPOCK1 on cell proliferation, migration and invasion assays of ESCC cells
To investigate the functional role of SPOCK1 in ESCC cells, we performed loss-of-function assays using si-SPOCK1-transfected ESCC cell lines. As shown in Figure 5a and b, SPOCK1 expression was effectively reduced in si-SPOCK1-transfected TE-8 and TE-1 cells. Functional assays showed that cell proliferation, migration and invasion were inhibited in si-SPOCK1-transfected cells compared with those in mock- or siRNA-control-transfected cells (P<0.0001; Figure 5c–e).
Loss-of-function studies using siRNAs. (a) Expression levels of SPOCK1 mRNA 72 h after transfection with si-SPOCK1 in ESCC cell lines. (b) SPOCK1 protein expression 72 h after transfection with si-SPOCK. (c) Cell proliferation was determined by XTT assays. Inhibition of cell proliferation was observed in si-SPOCK1-transfected cell lines. *P<0.0001; **P<0.05. (d) Cell migration activity. (e) Cell invasion was determined by Matrigel invasion assays. Inhibition of migration and invasion was observed in si-SPOCK1-transfected cell lines. *P<0.0001.
Analysis of SPOCK1/SPOCK1 expression in ESCC clinical specimens
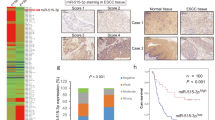
We validated the expression of SPOCK1 mRNA and protein in clinical ESCC specimens. Immunohistochemistry showed that SPOCK1 tended to be strongly expressed in the cytoplasm of ESCC lesions. Moderate or strong expression was observed in the nuclei of muscle layers, vessels and the basal layers of non-cancerous tissues (Figure 6a). SPOCK1 mRNA expression was upregulated in 20 ESCC specimens and cell lines compared with 10 non-cancerous tissues (P<0.05; Figure 6b).
Expression levels of SPOCK1 in ESCC clinical specimens. (a) Immunohistochemical staining of SPOCK1 in ESCC specimens (left panel, SPOCK1 staining; right panel, hematoxylin–eosin staining; original magnification, x100). (b) Expression levels of SPOCK1 mRNA in ESCC clinical specimens. GUSB was used as an internal control.
Discussion
Owing to local invasion and distant metastasis at the time of the initial diagnosis, ESCC is one of the most lethal digestive tract cancers in Japan.23 Molecularly targeted therapies for head and neck squamous cell carcinoma have shown remarkable therapeutic efficacy; however, no targeted therapeutics have been approved for treatment of ESCC.24 Therefore, new treatment options based on current genomic approaches are needed. Dysregulated miRNAs and miRNA-mediated RNA networks are deeply involved in ESCC pathogenesis. During the past few years, our research group (and others) reported that many miRNAs contribute to ESCC development, metastasis and drug resistance.10, 11, 21, 25
Based on miRNA expression signatures of cancers, we have identified antitumor miRNAs that modulate novel cancer networks in several cancers, including ESCC.26, 27 Current RNA sequencing technology can be used to illuminate the miRNA expression signatures of cancer cells.20, 27 We recently constructed RNA sequencing-based miRNA expression signatures using laryngeal and hypopharyngeal squamous cell carcinoma clinical specimens.20 These signatures revealed that several passenger strands of miRNAs were aberrantly expressed in cancer tissues.17, 19, 20 In this study, we demonstrated that the passenger strand of miR-150-3p acted as an antitumor miRNA as well as the guide strand of miR-150-5p in ESCC cells. Past studies showed that miR-150-5p was a multifunction miRNA with both oncogenic and antitumor functions depending on the cancer type.28, 29
In ESCC cells, miR-150-5p directly regulated ZEB1 as an inducer of the epithelial–mesenchymal transition.30 Moreover, the expression status of miR-150-5p was associated with clinicopathological features of patients with ESCC.30 Another study showed that overexpression of miR-150-5p suppressed ESCC cell proliferation i n vitro and in vivo through targeting of 5S ribosomal RNA.31 These findings indicated that miR-150-5p acted as an antitumor miRNA in ESCC cells. In contrast, the functional roles of the passenger strand of miR-150-3p in ESCC have been unclear. Our present data are the first to show that low expression of miR-150-3p in ESCC clinical specimens and ectopic expression of the miRNA significantly inhibited ESCC cell migration and invasion.
More recently, we showed that dual strands of pre-miR-150 acted as antitumor miRNAs in HNSCC cells.20 Interestingly, fivve putative candidate genes involved in the focal adhesion pathway (TNC, ITGA3, ITGA6, CAV2 and XIAP) were regulated by both miR-150-5p and miR-150-3p in HNSCC cells.20 In miRNA studies, one of the main challenges is the identification of target genes in cancer cells. In this study, a total of 12 genes were found to be putative targets of both miR-150-5p and miR-150-3p regulation in ESCC cells.
Our present data showed that SPOCK1 was directly regulated by two antitumor miRNAs, miR-150-5p and miR-150-3p, in ESCC cells. Functional assays showed that silencing of SPOCK1 in ESCC cells significantly inhibited cancer cell migration and invasion. Overexpression of SPOCK1 has been observed in several cancers and SPOCK1 has a central role in cancer cell progression, metastasis and drug resistance.32, 33, 34 In lung cancer, expression levels of SPOCK1 were significantly higher in metastatic cancer tissues than in non-metastatic tissues.35 Silencing of SPOCK1 expression in lung cancer cells suppressed colony formation and invasion.35 Moreover, SPOCK1 expression was induced by tumor growth factor-β, and SPOCK1 promoted epithelial–mesenchymal transition marker expression.35 In ESCC cells, overexpression of SPOCK1 in low malignancy Eca109 cells induced epithelial–mesenchymal transition phenotypes.36 Similarly, in ESCC cells, overexpression of SPOCK1 in RWPE-1 non-malignant prostatic epithelial cell enhanced cell migration and invasion abilities.37 Taken together, overexpression of SPOCK1 is involved in ESCC pathogenesis, and SPOCK1-mediated cancer pathways might be a putative therapeutic target in ESCC cells.
In conclusion, dual strands of pre-miR-150 (miR-150-5p and miR-150-3p) acted as antitumor miRNAs in ESCC cells. SPOCK1 was directly regulated by both miR-150-5p and miR-150-3p and overexpression of SPOCK1 enhanced ESCC cell aggressiveness. The involvement of passenger strand miRNAs in the regulation of cancer cell aggressiveness is a novel concept in RNA research. Elucidation of novel pre-miR-150/SPOCK1-mediated molecular pathways may lead to a better understanding ESCC pathogenesis.
References
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015).
Hori, M. M. T., Shibata, A., Katanoda, K., Sobue, T., Nishimoto, H. et al. Cancer incidence and incidence rates in Japan in 2012: a study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J. Clin. Oncol. 45, 884–891 (2016).
Pennathur, A., Gibson, M. K., Jobe, B. A. & Luketich, J. D. Oesophageal carcinoma. Lancet 381, 400–412 (2013).
Enzinger, P. C. & Mayer, R. J. Esophageal cancer. N. Engl. J. Med. 349, 2241–2252 (2003).
Ohashi, S., Miyamoto, S., Kikuchi, O., Goto, T., Amanuma, Y. & Muto, M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology 149, 1700–1715 (2015).
Hongo, M., Nagasaki, Y. & Shoji, T. Epidemiology of esophageal cancer: orient to occident. Effects of chronology, geography and ethnicity. J. Gastroenterol. Hepatol. 24, 729–735 (2009).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Isozaki, Y., Hoshino, I., Nohata, N., Kinoshita, T., Akutsu, Y., Hanari, N. et al. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int. J. Oncol. 41, 985–994 (2012).
Osako, Y., Seki, N., Kita, Y., Yonemori, K., Koshizuka, K., Kurozumi, A. et al. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int. J. Oncol. 49, 2255–2264 (2016).
Harada, K., Baba, Y., Ishimoto, T., Shigaki, H., Kosumi, K., Yoshida, N. et al. The role of microRNA in esophageal squamous cell carcinoma. J. Gastroenterol. 51, 520–530 (2016).
Kikkawa, N., Hanazawa, T., Fujimura, L., Nohata, N., Suzuki, H., Chazono, H. et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). Br. J. Cancer 103, 877–884 (2010).
Nohata, N., Hanazawa, T., Kikkawa, N., Sakurai, D., Fujimura, L., Chiyomaru, T. et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br. J. Cancer 105, 833–841 (2011).
Itesako, T., Seki, N., Yoshino, H., Chiyomaru, T., Yamasaki, T., Hidaka, H. et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS ONE 9, e84311 (2014).
Fukumoto, I., Hanazawa, T., Kinoshita, T., Kikkawa, N., Koshizuka, K., Goto, Y. et al. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer 112, 891–900 (2015).
Matsushita, R., Seki, N., Chiyomaru, T., Inoguchi, S., Ishihara, T., Goto, Y. et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br. J. Cancer 113, 282–289 (2015).
Matsushita, R., Yoshino, H., Enokida, H., Goto, Y., Miyamoto, K., Yonemori, M. et al. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressiveness. Oncotarget 7, 28460–28487 (2016).
Mataki, H., Seki, N., Mizuno, K., Nohata, N., Kamikawaji, K., Kumamoto, T. et al. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 7, 72084–72098 (2016).
Koshizuka, K., Nohata, N., Hanazawa, T., Kikkawa, N., Arai, T., Okato, A. et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget 8, 30288–30304 (2017).
Kano, M., Seki, N., Kikkawa, N., Fujimura, L., Hoshino, I., Akutsu, Y. et al. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 127, 2804–2814 (2010).
Kita, Y., Nishizono, Y., Okumura, H., Uchikado, Y., Sasaki, K., Matsumoto, M. et al. Clinical and biological impact of cyclin-dependent kinase subunit 2 in esophageal squamous cell carcinoma. Oncol. Rep. 31, 1986–1992 (2014).
Berry, M. F. Esophageal cancer: staging system and guidelines for staging and treatment. J. Thorac. Dis. 6 (Suppl 3), S289–S297 (2014).
Liu, Y., Xiong, Z., Beasley, A., D'Amico, T. & Chen, X. L. Personalized and targeted therapy of esophageal squamous cell carcinoma: an update. Ann. NY. Acad. Sci. 1381, 66–73 (2016).
Koumangoye, R. B., Andl, T., Taubenslag, K. J., Zilberman, S. T., Taylor, C. J., Loomans, H. A. et al. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol. Cancer 14, 24 (2015).
Mizuno, K., Mataki, H., Arai, T., Okato, A., Kamikawaji, K., Kumamoto, T. et al. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J. Hum. Genet. (e-pub ahead of print 9 March 2017; doi: 10.1038/jhg.2017.27).
Goto, Y., Kojima, S., Nishikawa, R., Kurozumi, A., Kato, M., Enokida, H. et al. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 113, 1055–1065 (2015).
Li, J., Hu, L., Tian, C., Lu, F., Wu, J. & Liu, L. microRNA-150 promotes cervical cancer cell growth and survival by targeting FOXO4. BMC. Mol. Biol. 16, 24 (2015).
Qu, Y., Pan, S., Kang, M., Dong, R. & Zhao, J. MicroRNA-150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biol. 37, 5275–5284 (2015).
Yokobori, T., Suzuki, S., Tanaka, N., Inose, T., Sohda, M., Sano, A. et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 104, 48–54 (2013).
Wang, X., Ren, Y., Wang, Z., Xiong, X., Han, S., Pan, W. et al. Down-regulation of 5 S rRNA by miR-150 and miR-383 enhances c-Myc-rpL11 interaction and inhibits proliferation of esophageal squamous carcinoma cells. FEBS Lett. 589 (Part B), 3989–3997 (2015).
Fan, L. C., Jeng, Y. M., Lu, Y. T. & Lien, H. C. SPOCK1 is a novel transforming growth factor-beta-induced myoepithelial marker that enhances invasion and correlates with poor prognosis in breast cancer. PLoS ONE 11, e0162933 (2016).
Li, Y., Chen, L., Chan, T. H., Liu, M., Kong, K. L., Qiu, J. L. et al. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology 144, 179–191 and 194 (2013).
Ma, L. J., Wu, W. J., Wang, Y. H., Wu, T. F., Liang, P. I., Chang, I. W. et al. SPOCK1 overexpression confers a poor prognosis in urothelial carcinoma. J. Cancer 7, 467–476 (2016).
Miao, L., Wang, Y., Xia, H., Yao, C., Cai, H. & Song, Y. SPOCK1 is a novel transforming growth factor-beta target gene that regulates lung cancer cell epithelial–mesenchymal transition. Biochem. Biophys. Res. Commun. 440, 792–797 (2013).
Song, X., Han, P., Liu, J., Wang, Y., Li, D., He, J. et al. Up-regulation of SPOCK1 induces epithelial–mesenchymal transition and promotes migration and invasion in esophageal squamous cell carcinoma. J. Mol. Histol. 46, 347–356 (2015).
Chen, Q., Yao, Y. T., Xu, H., Chen, Y. B., Gu, M., Cai, Z. K. et al. SPOCK1 promotes tumor growth and metastasis in human prostate cancer. Drug Des. Dev. Ther. 10, 2311–2321 (2016).
Acknowledgements
This study was supported by KAKENHI Grants 15K10801(C), 15K10108 and 17H04285(B). We wish to thank the Joint Research Laboratory, Kagoshima University Graduate School of Medical and Dental Sciences, for the use of their facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Osako, Y., Seki, N., Koshizuka, K. et al. Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer cell migration and invasion in esophageal squamous cell carcinoma. J Hum Genet 62, 935–944 (2017). https://doi.org/10.1038/jhg.2017.69
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.69
This article is cited by
-
Urinary microRNA biomarkers for detecting the presence of esophageal cancer
Scientific Reports (2021)
-
Molecular pathogenesis of breast cancer: impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes
Journal of Human Genetics (2021)
-
RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation
Journal of Human Genetics (2020)
-
Regulation of aberrantly expressed SERPINH1 by antitumor miR-148a-5p inhibits cancer cell aggressiveness in gastric cancer
Journal of Human Genetics (2020)
-
Gene regulation by antitumor miR-130b-5p in pancreatic ductal adenocarcinoma: the clinical significance of oncogenic EPS8
Journal of Human Genetics (2019)