Abstract
The aim of this study was to investigate the parthenogenetic origin of fetiform teratoma by using molecular genetic studies and methylation status analyses. A fetiform teratoma was removed from a 35-year-old nulligravida woman. Genotyping of microsatellite marker loci, microarray analysis of single-nucleotide polymorphism (SNP) loci and methylation status analysis of the differentially methylated region (DMR) within the human IGF2-H19 locus were performed. Karyotypes of the host and the fetiform teratoma were 46, XX. The fetiform teratoma was homozygous at all loci and meiotic recombinations in the tumor were confirmed by SNP microarray analysis. Methylation analysis indicated that the host had both methylated and unmethylated IGF2-H19 DMR alleles, while the fetiform teratoma had unmethylated alleles only. Genetically, the fetiform teratoma had homozygous genotypes with meiotic recombination and a duplicated unmethylated host allele, indicating that it was a parthenogenetic tumor arising from a mature ovum after meiosis II. This is the first demonstration of a fetiform teratoma originating from a mature haploid ovum.
Similar content being viewed by others
Introduction
Fetiform teratoma is a very rare form of ovarian mature cystic teratoma that resembles a malformed fetus.1, 2, 3, 4, 5, 6, 7, 8 The origin and methylation status of fetiform teratoma have yet to be determined.7 Ovarian mature cystic teratomas are composed of fully differentiated mature tissues derived from all three germ layers (ectodermal, endodermal and mesodermal), although these components are generally disorganized. They contain a cyst filled with sebaceous material, desquamated squamous cells and hair. Previously, using genotype analysis of microsatellite marker loci, we showed that the origin of each was at a different stage of female gametogenesis (oogenesis) among 25 ovarian mature cystic teratomas.9 It is now thought that ovarian mature cystic teratomas are parthenogenetic tumors that can arise at all stages of oogenesis.9, 10, 11, 12, 13 However, no case had been described that arose from a mature ovum after meiosis II, in which genotypes at all loci were homozygous with alleles inherited from the mother and the father due to meiotic recombination.9
Normally, in the differentially methylated region (DMR) within the human IGF2-H19 locus, the paternal allele is methylated and the maternal allele is unmethylated.14 The degree of H19 hypomethylation was found to increase as the oogenesis stage of origin of the mature cystic teratomas advanced.9 Therefore, the DMR within the human IGF2-H19 locus is unmethylated in mature ova. However, in our previous study, among 25 mature ovarian cystic teratomas, no cases had an unmethylated status at the DMR within IGF2-H19.9
We recently encountered for the first time a case of fetiform teratoma that originated from a mature ovum. Here, we report the results of molecular genetic studies and methylation status analyses of this teratoma.
Materials and methods
A case of fetiform teratoma
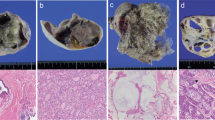
A 35-year-old nulligravida woman was referred to Niigata City General Hospital for ultrasonographic diagnosis of a right side adnexal mass. She had a regular menstrual period. There was no important notice in her past history and family history. Ultrasonographic imaging of the pelvis revealed a complex cystic mass within the right adnexa that measured 10 × 8 cm. X-radiography showed ossification in the pelvic cavity (Figure 1a). Computed tomography showed a right side ovarian tumor (10 × 8 cm) that included fat and ossified material (Figure 1b). This composition was confirmed by magnetic resonance imaging. In addition, the serum levels of CA125 and squamous cell carcinoma antigen were within normal ranges (<35 U ml−1 and <1.5 ng ml−1, respectively). Based on these findings, the preoperative diagnosis was mature ovarian cystic teratoma of the right ovary.
Clinical findings of the fetiform teratoma. (a) Transvaginal ultrasonographic examination before surgery. A complex cystic mass (10 × 8 cm) was seen in the right adnexal region. (b) Computed tomography examination before surgery. Right side ovarian tumor included fat (*) and ossified material (**). (c) Front view of the fetiform teratoma. The fetiform mass was attached to the tumor capsule by its cranial surface. The fetiform mass had both upper and lower extremities. A phallus-like structure (arrow) and pubic hair were seen in the lower trunk. (d) Posterior view of the fetiform mass. The fetiform mass had both upper and lower extremities, hips and buttocks. The whole body was covered with fine lanugo, and its skin was generally thick and hard. (e) X-radiography of the fetiform mass. Ossification of the cranium floor, some cervical vertebrae and both scapulae was detected. A full color version of this figure is available at the Journal of Human Genetics Journal online.
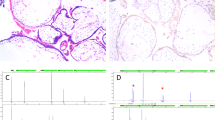
Surgical intervention consisted of a right salpingo-oophorectomy. When the yellow sebaceous material was removed, a doll-like structure (8 cm in height) appeared (Figures 1c and d). The fetiform mass was attached to the tumor capsule by its cranial surface. Figure 1c shows a front view of the fetiform mass that had both upper and lower extremities with hips and buttocks. A phallus-like structure and pubic hair were seen in the lower trunk. Figure 1d shows a posterior view of the fetiform mass. The whole body was covered with fine lanugo, and its skin was generally thick and hard. X-radiography of the fetiform mass showed ossification of the cranium floor, some cervical vertebrae, and left and right scapulae (Figure 1e). Pathological examination showed that the fetiform mass was covered with squamous epithelium, and included smooth muscle, fat, bone and urethra tissues. Based on the above findings, this ovarian tumor was diagnosed as a fetiform teratoma. All samples were obtained after receiving written informed consent, and our study protocol was approved by the Institutional Review Board for Ethical, Legal and Social Issues of Nagasaki University (No. 12121775). All experiments in this study were performed in accordance with the appropriate guidelines.
Genotyping at microsatellite marker loci
Whole-blood samples were obtained from the patient (host). Genomic DNA was extracted from the host’s white blood cells, and tissue samples were taken from the upper trunk (FT-1) and the lower extremities (FT-2) of the teratoma. Host, FT-1 and FT-2 were genotyped at 16 microsatellite markers using the AmpFISTR Identifiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). DNA was amplified by PCR and PCR products were analyzed using a 3100 Genetic Analyzer (Applied Biosystems).
Copy number analysis and genotyping of SNP loci
We performed genome-wide SNP genotyping using the Genome-Wide Human SNP Array 6.0 (SNP6.0), following the manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA). Genotyping of the samples was performed using the Genotyping Console (GTC) 4.0. We analyzed copy number change, loss of heterozygosity and allelic ratios for the fetiform teratoma using Partek Genomics Suite, version 6.5 (Partek Inc., St Louis, MO, USA). The genomic positions of SNPs corresponded to those in the March 2006 human genome release (hg18). Copy number estimates for the fetiform teratoma were achieved using reference data from 80 normal control samples.
To determine the haplotypes of the host and her fetiform teratoma, we performed genome-wide SNP genotyping in the host, her parents and the fetiform teratoma, using Cytoscan HD array, following the manufacturer’s instructions (Affymetrix). Out of 749 157 SNP loci, 35 390 SNPs were selected as informative loci (discordantly homozygous between the host’s parents (AA and BB), while the genotype of the host was heterozygous (AB)). At all of the informative loci, as it was confirmed that the genotypes of fetiform teratoma were homozygous, it was determined that meiotic recombination had occurred given the presence of mixed haplotypes derived from the host’s parents on the same chromosome.
Methylation assay at the IGF2-H19 locus
The fetiform teratoma was analyzed for methylation status of the DMR at the human IGF2-H19 locus on chromosome 11p15.5. To differentiate between host and fetiform teratoma alleles, we assayed the 8097A/G SNP (Genbank Accession No. AF125183, rs2107425) within the DMR.15 Primer sequences were as follows: forward, 5′-GGACGGAATTGGTTGTAGTT-3′; reverse, 5′-AGGCAATTGTCAGTTCAGTAA-3′.16 PCR was performed using the GeneAmp PCR system 9700 (Applied Biosystems) with cycling conditions of 2 min at 95 °C; 35 cycles of 1 min at 95 °C, 20 s at 56 °C and 20 s at 72 °C; and one cycle of 10 min at 72 °C. PCR products were analyzed by agarose gel electrophoresis and sequenced on a 3100 Genetic Analyzer (Applied Biosystems).
DNA was modified with sodium bisulfite using the CpGenome DNA Modification Kit (Chemicon International, Temicula, CA, USA), as per the manufacturer’s instructions.17 After the conversion of DNA, the methylation status of the DMR was examined. Methylation-specific PCR was performed in a 10-μl mixture containing 1 μl of modified DNA, 1 μl of 10 × PCR buffer, 0.8 μl of GeneAmp dNTP Mix (Applied Biosystems), 1 μl of each of the forward and reverse primers, and 0.05 μl of AmpliTaq Gold (Applied Biosystems). Cycling conditions were initial denaturation at 95 °C for 12 min, 60 cycles of 95 °C for 45 s and 55 °C (or 49 °C) for 20 s, and final extension at 72 °C for 20 s. The sequences of the primers, designed by Poon et al.,16 were as follows: methylated allele-specific forward primer (M-for), 5′-TTAATTGGGGTTCGTTCG-3′; methylated allele-specific reverse primer (M-rev), 5′-CCCGACCTAAAAATCTAATACGA-3′; unmethylated allele-specific forward primer (U-for), 5′-GGTTTGTTTGTGGAAATGTTTT-3′; and unmethylated allele-specific reverse primer (U-rev), 5′-CCCAACCTAAAAATCTAATACAA-3′. PCR products were cloned into 2.1 TOPO using the TOPO TA Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced on a 3100 Genetic Analyzer.
Results
Genotypes of host and fetiform teratoma (FT-1 and FT-2)
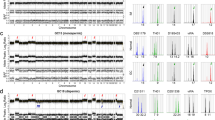
Sixteen microsatellite markers were genotyped in the host and samples of her fetiform teratoma (FT-1 and FT-2). Of these, nine were informative. The host was heterozygous at all nine loci, while the genotypes of FT-1 and FT-2 were identically homozygous at all loci including the amelogenin locus (Table 1). In addition, both FT-1 and FT-2 also inherited one single-nucleotide polymorphism (SNP) allele from the host, and their genotypes at all loci were identical (Figures 2a and b). Upon comparison with DNA microarray data of a male with a normal karyotype, the copy numbers of all SNP loci in the fetiform teratoma were identical to the control for the autosomal chromosomes, and were double for the X chromosome (Figure 2c). Based on these findings, the genome volume in the fetiform teratoma was equivalent to that of a normal female karyotype (46,XX).
SNP6.0 array plots for the fetiform teratoma and genome-wide SNP copy number in the fetiform teratoma. (a) SNP6.0 array plots for chromosome 1. (b) SNP6.0 array plots for the X chromosome. Both homozygous and heterozygous SNP loci were observed in the host. Only homozygous SNP loci were observed in FT-1 and FT-2, the genotypes being concordant at all loci. (c) Genome-wide SNP copy number in the fetiform teratoma. SNP copy number in the fetiform teratoma was analyzed with respect to the genome volume of a male with the 46,XY karyotype, which included two copies. For chromosomes 1 to 22, the copy numbers of all SNP loci in the fetiform teratoma were identical to those in a male with the normal karyotype (46,XY). The copy numbers of X chromosome SNP loci in the fetiform teratoma were double those in a male with the normal karyotype.
Haplotypes of the host and fetiform teratoma
Heterozygous loci (AB genotype) in the host, derived from the AA genotype in the host’s father and the BB genotype in the host’s mother, were informative to analyze the haplotype in the fetiform teratoma; this enabled the origins of the chromosomal segments to be confirmed. Out of 749 157 SNP loci, 35 390 SNPs were selected as informative loci (discordantly homozygous between the host’s parents (AA and BB), while the genotype of the host was heterozygous (AB)). The fetiform teratoma had complete homozygosity at all informative loci. In multiple chromosomes, fetiform teratoma had patchy chromosomal segments, which showed the presence of mixed SNP domains derived from the host’s father and the host’s mother on the same chromosome (Figure 3).
Haplotype origins of the fetiform teratoma. Heterozygous loci (AB genotype) in the host, derived from the AA genotype in the host’s father and the BB genotype in her mother, were analyzed; the origins of chromosomal segments were confirmed in fetiform teratoma by haplotype analyses using Cytoscan HD microarray. Chromosomal segments in red hatching were derived from the host’s mother and segments without red hatching were from her father. A full color version of this figure is available at the Journal of Human Genetics Journal online.
Methylation status of IGF2-H19 DMR in the host and fetiform teratoma
DMR-SNP analysis showed that the host genotype was GA heterozygous, while that of FT-1 and FT-2 was AA homozygous (Table 2). The methylation status of the DMR at the IGF2-H19 locus was assessed by bisulfite treatment, and methylated or unmethylated sequence-specific PCR, followed by cloning of the PCR products. Normally, only the paternal allele in the DMR is methylated, while the maternal allele remains unmethylated.14 Both methylated and unmethylated alleles were amplified in the host, while only the unmethylated allele was amplified in FT-1 and FT-2 (Figure 4). In the host, all 10 clones (100%) showing methylated patterns had the G allele and all 10 clones (100%) showing unmethylated patterns had the A allele (Table 2). In both FT-1 and FT-2, of the 10 clones with the A allele, all showed an unmethylated pattern; none showed a methylated pattern (Table 2). The methylation patterns of the host and the fetiform teratoma were thus different.
Methylation-specific PCR in the fetiform teratoma. M-primer, methylated primer; U-primer, unmethylated primer; Host, patient blood DNA; FT-1, DNA from upper trunk of the fetiform teratoma; FT-2, DNA from a lower extremity of the fetiform teratoma. Both methylated and unmethylated DMR IGF2-H19 alleles were amplified in the host, while only the unmethylated allele was observed in FT-1 and FT-2. A full color version of this figure is available at the Journal of Human Genetics Journal online.
Discussion
Fetiform teratoma is a rare form of ovarian mature cystic teratoma that is highly developed and organized, resembling a fetus-like structure.1, 2, 3, 4, 5, 6, 7, 8 In this study, for the first time, we performed molecular genetic studies on the origin and methylation status of a fetiform teratoma to help understand its pathogenesis.
When a fetus-like mass is observed, it is necessary to perform a differential diagnosis among fetiform teratoma, fetus in fetu and ectopic pregnancy.6, 7, 8 It has been proposed that fetiform teratoma and fetus in fetu can be differentiated based on zygosity.6, 7, 8, 15 The fetiform teratoma in this case was homozygous at all loci. Fetus in fetu is genetically identical to its host; the results obtained in this case were thus consistent with fetiform teratoma and fetus in fetu could be ruled out. However, fetiform teratomas must also be distinguished from ectopic pregnancies. For this, ectopic pregnancies generally include villous tissues, while fetiform ovarian teratomas do not. Furthermore, ectopic pregnancies, which are fertilized products, have both paternal and maternal alleles; therefore, the genotypes should have both heterozygous and homozygous loci. The fetiform teratoma examined here showed homozygosity at all loci and had only alleles of maternal (host) origin and pathological examination showed an absence of villous components. This ruled out the possibility of an ectopic pregnancy.
Previously, we established the germ cell origin of ovarian mature cystic teratomas using features such as the homozygosity of genetic polymorphisms that are heterozygous in the hosts, and crossover events.9 Among 25 mature cystic ovarian teratomas, it was confirmed that they originated from each stage of oogenesis (groups Ia, IIa, Ib and IIb), but not from the mature ovum stage.9 Group Ia teratomas, with heterozygosity at pericentromeric loci and no chromosomal recombination, originated from premeiotically dividing germ cells; group IIa teratomas, with homozygosity at pericentromeric loci and no chromosomal recombination, originated from apparently immature primary oocytes; group Ib teratomas, with heterozygosity at pericentromeric loci and chromosomal recombination, originated from primary oocytes undergoing meiotic recombination; while group IIb teratomas, with homozygosity at pericentromeric loci and chromosomal recombination, arose by an error in meiosis II after the completion of meiosis I. Ovarian teratomas, which originate from mature ova after the completion of meiosis II, should have complete homozygosity at all loci as well as chromosomal recombination. In the case of complete homozygosity at all loci, a group IIa teratoma or teratoma from mature ovum is possible. The only way to discriminate group IIa teratoma and teratoma from mature ovum is to confirm the presence of chromosomal recombination in ovarian teratoma. In this study, we genotyped the host’s parents to determine the haplotypes of the host and her fetiform teratoma. Heterozygous SNP loci in the host, at which the host’s parents had discordant homozygous SNPs, were selected as informative loci (discordantly homozygous between the host’s parents (AA and BB), while the genotype of the host was heterozygous (AB)). At all of the informative loci, as it was confirmed that the genotypes of fetiform teratoma were homozygous, it was determined that meiotic recombination had occurred given the presence of mixed haplotypes derived from the host’s parents on the same chromosome. In multiple chromosomes of fetiform teratoma, patchy haplotypes, which included SNP domains derived from both the host’s father and her mother, were confirmed to be in a homozygous state (Figure 3). Therefore, the fetiform teratoma was confirmed to have complete homozygosity at all loci and have arisen from a mature ovum after the completion of chromosomal meiotic recombination.
The methylation status of the DMR at the IGF2-H19 locus was heterozygous in the host, with a methylated G allele and an unmethylated A allele. On the other hand, the fetiform teratoma genotype was homozygous for the unmethylated A allele. The methylation status of the human IGF2-H19 DMR is useful because the methylated allele is of paternal origin and the unmethylated allele is of maternal origin in human tissues.14 This enables us to determine the stage of oogenesis from which the teratoma originated. The degree of methylation at human IGF2-H19 DMR progressively decreases as the presumptive origin of the tumors advances through the stages of oogenesis,9 suggesting that ovarian teratomas originating from mature ova would have an unmethylated status. In our previous study, group IIa teratomas showed homozygosity defined by a consistent difference in allele intensity between the tumor and host, and demonstrated partial methylation at human IGF2-H19 DMR.9 Meanwhile, the present fetiform teratoma showed an unmethylated status at human IGF2-H19 DMR. Therefore, based on the patchy homozygous haplotype and methylation status, we demonstrated that the fetiform teratoma was a parthenogenetic tumor arising from a mature ovum after the completion of meiosis II. To our knowledge, this is the first report of its kind. Notably, skeletal muscle was not identified in previously reported cases of fetiform teratoma, and was not found in the present case either, suggesting that the paternal genome may be required for this morphogenesis.6
In conclusion, we have shown a fetiform ovarian teratoma to be of parthenogenetic origin, arising from a mature ovum after the completion of meiosis II. We were able to determine the presence of different methylation patterns between host and fetiform teratoma. These findings should contribute to future studies on the pathogenesis of fetiform teratoma.
References
Weldon-Linne, C. M. & Rushovich, A. M. Benign ovarian cystic teratomas with homunculi. Obstet. Gynecol. 61, 88S–94S (1983).
Abbott, T. M., Hermann, W. J. Jr & Scully, R. E. Ovarian fetiform teratoma (homunculus) in a 9-year-old girl. Int. J. Gynecol. Pathol. 2, 392–402 (1984).
Inoue, M., Mitsuda, N., Tanaka, Y., Suehara, N., Ueda, G. & Kurachi, K. Benign cystic teratoma of ovary containing a homunculus. Nihon Sanka Fujinka Gakkai Zasshi 37, 1050–1053 (1985).
Miyake, J. & Ireland, K. Ovarian mature teratoma with homunculus coexisting with an intrauterine pregnancy. Arch. Pathol. Lab. Med. 110, 1192–1194 (1986).
Lee, Y. H., Kim, S. G., Choi, S. H., Kim, I. S. & Kim, S. H. Ovarian mature cystic teratoma containing homunculus: a case report. J. Korean Med. Sci. 18, 905–907 (2003).
Kuno, N., Kadomatsu, K., Nakamura, M., Miwa-Fukuchi, T., Hirabayashi, N. & Ishizuka, T. Mature ovarian cystic teratoma with a highly differentiated homunculus: a case report. Birth Defects Res. A Clin. Mol. Teratol. 70, 40–46 (2004).
Weiss, J. R., Burgess, J. R. & Kaplan, K. J. Fetiform teratoma (homunculus). Arch. Pathol. Lab. Med. 130, 1552–1556 (2006).
Greenberg, J. A. & Clancy, T. E. Fetiform teratoma (homunculus). Rev. Obstet. Gynecol. 1, 95–96 (2008).
Miura, K., Obama, M., Yun, K., Masuzaki, H., Ikeda, Y., Yoshimura, S. et al. Methylation imprinting of H19 and SNRPN genes in human benign ovarian teratomas. Am. J. Hum. Genet. 65, 1359–1367 (1999).
Linder, D., McCaw, B. K. & Hecht, F. Parthenogenic origin of benign ovarian teratomas. N. Engl. J. Med. 292, 63–66 (1975).
Dahl, N., Gustavson, K. H., Rune, C., Gustavsson, I. & Pettersson, U. Benign ovarian teratomas. An analysis of their cellular origin. Cancer Genet. Cytogenet. 46, 115–123 (1990).
Deka, R., Chakravarti, A., Surti, U., Hauselman, E., Reefer, J., Majumder, P. P. et al. Genetics and biology of human ovarian teratomas. II. Molecular analysis of origin of nondisjunction and gene-centromere mapping of chromosome I markers. Am. J. Hum. Genet. 47, 644–655 (1990).
Surti, U., Hoffner, L., Chakravarti, A. & Ferrell, R. E. Genetics and biology of human ovarian teratomas. I. Cytogenetic analysis and mechanism of origin. Am. J. Hum. Genet. 47, 635–643 (1990).
Bartolomei, M. S. & Tilghman, S. M. Genomic imprinting in mammals. Annu. Rev. Genet. 31, 493–525 (1997).
Miura, S., Miura, K., Yamamoto, T., Yamanaka, M., Saito, K., Hirabuki, T. et al. Origin and mechanisms of formation of fetus-in-fetu: two cases with genotype and methylation analyses. Am. J. Med. Genet. A 140, 1737–1743 (2006).
Poon, L. L., Leung, T. N., Lau, T. K., Chow, K. C. & Lo, Y. M. Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clin. Chem. 48, 35–41 (2002).
Kubota, T., Das, S., Christian, S. L., Baylin, S. B., Herman, J. G. & Ledbetter, D. H. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 16, 16–17 (1997).
Acknowledgements
We would like to thank Yasuko Noguchi for her technical contribution. KM and HM were supported by the Japan Society for the Promotion of Science KAKENHI (grant numbers 26462495 and 25462563, respectively).
Author contributions
KM wrote the main manuscript text, performed genotyping analysis of microsatellite markers and methylation analysis and prepared Figure 4. TK and TI were the doctors attending this case of fetiform teratoma and prepared Figure 1. CS, KS and K-iY performed SNP microarray genotyping analysis and prepared Figures 2 and 3. HM interpreted the data, performed critical revisions of the manuscript and supervised the study. All authors reviewed the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Miura, K., Kurabayashi, T., Satoh, C. et al. Fetiform teratoma was a parthenogenetic tumor arising from a mature ovum. J Hum Genet 62, 803–808 (2017). https://doi.org/10.1038/jhg.2017.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.45