Abstract
Our studies of microRNA (miRNA) expression signatures have shown that microRNA-1 (miR-1) and microRNA-206 (miR-206) were downregulated in head and neck squamous cell carcinoma (HNSCC) clinical specimens. The seed sequences of these miRNAs are identical, suggesting that the identification of the molecular targets regulated by miR-1 and miR-206 will provide new insights into novel mechanisms of HNSCC pathogenesis. Our present data showed that restoration of miR-1 and miR-206 significantly inhibited HNSCC cells’ aggressiveness. A combination of gene expression data and in silico analysis revealed that several pathways (‘pathway in cancer’, ‘focal adhesion pathway’, ‘MAPK signaling pathway’, ‘regulation of actin cytoskeleton pathway’ and ‘ECM–receptor interaction pathway’) were regulated by miR-1 and miR-206. Among them, we found that two growth factor receptors, epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (c-MET), were directly regulated by both miR-1 and miR-206 in HNSCC cells. Also, downstream oncogenic signaling of these receptors was reduced by restoration of miR-1 or miR-206 expression. Moreover, overexpression of EGFR and c-MET was observed in HNSCC clinical specimens. The identification of targets modulated by tumor-suppressive miR-1 and miR-206 may lead to a better understanding of molecular pathogenesis of HNSCC.
Similar content being viewed by others
Introduction
The 5-year survival frequency of patients with head and neck squamous cell carcinoma (HNSCC) is ~50% because such patients are usually diagnosed at a late stage.1 Moreover, local tumor recurrence and distant metastasis appear after conventional therapies.1 Application of genomic approaches might elucidate novel molecular pathways underlying HNSCC and thereby improve therapeutic approaches to the disease.
The discovery of noncoding (NC) RNAs in the human genome was an important conceptual breakthrough for cancer research in the post-genome sequencing era.2 NCRNAs affect every stage of gene expression, from gene transcription and translation to messenger RNA (mRNA) stability.3, 4 Among NCRNAs, the microRNAs (miRNAs) are small NCRNA molecules (18–25 nucleotides in length) that regulate the expression of protein-coding/non-protein-coding genes by repressing translation or cleaving RNA transcripts in a sequence-specific manner.5 Accumulating evidence has demonstrated pivotal roles for miRNAs in human cancer pathogenesis.6, 7 Several studies have shown that hyperactivity or diminished function of microRNAs disrupt the tightly controlled RNA networks in cancer cells.4, 6, 7 Dysregulated RNA networks contribute to the development of cancer cells and the acquisition of the malignant phenotype.
We hypothesize that the identification of aberrantly expressed miRNA would be an important first step toward elucidating the details of miRNA-mediated oncogenic pathways. On the basis of this theory, we have constructed miRNA expression signatures of HNSCC clinical specimens and identified aberrantly expressed miRNAs.8, 9, 10, 11 Our previous studies showed that the miR-29 family (miR-29a/b/c), miR-218, miR-451a and the miR-26 family (miR-26a/b) were significantly reduced in HNSCC tissues. Furthermore, ectopic expression of these miRNAs inhibited cancer cell aggressiveness through the targeting of genes involved in ‘focal adhesion’ and ‘extracellular matrix (ECM)–receptor interaction’ pathways.8, 9, 12, 13, 14
For example, aberrant expression of ECM components and activation of ECM-mediated signals have been observed in cancer lesions and are known to trigger cancer cell aggressiveness.15, 16, 17 We demonstrated that laminin-332-integrin α6β4 signaling contribute to cancer cell migration and invasion in HNSCC, and these ECM components were regulated by the miR-29 family as well as miR-218.13, 14 Moreover, our recent study showed that downregulation of miR-223 enhanced ITGA3/ITGB1 signaling and contributed to cancer cell migration and invasion by prostate cancer cells.17 These data suggested that identification of miRNA-mediated oncogenic signaling pathways may have applications in the development of novel therapies targeted against metastatic cancers.
In this study, we focused on miR-1 and miR-206 because the expression of these miRNAs was downregulated in HNSCC signatures and the seed sequences of these miRNAs are identical.9, 18 The aim of this study was to investigate the functional significance of miR-1 and miR-206 in HNSCC and to identify novel molecular targets involved in HNSCC aggressiveness. Our present data showed that genes coding for epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (c-MET) were directly downregulated by both miR-1 and miR-206. Moreover, EGFR or c-MET were overexpressed in HNSCC clinical specimens. Targets regulated by tumor-suppressor miR-1 and miR-206 might provide important insights into the molecular mechanisms of HNSCC progression and metastasis and suggest novel therapeutic strategies for the treatment of the disease.
Materials and methods
Clinical HNSCC specimens
A total of 22 pairs of primary tumors and corresponding normal epithelial specimens and 23 formalin-fixed paraffin-embedded tissues were obtained from patients with HNSCC at Chiba University Hospital (Chiba, Japan) from 2008 to 2014. The patients’ backgrounds and clinicopathological characteristics are summarized in Tables 1 and 2. The patients were classified according to the 2002 Union for International Cancer Control (UICC) TNM staging criteria before treatment. Written consent for tissue donation for research purposes was obtained from each patient before tissue collection. The protocol was approved by the institutional review board of Chiba University.
The fresh specimens were immediately immersed in RNAlater (Qiagen, Valencia, CA, USA) and stored at −20 °C until RNA was extracted. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Cell line and cell culture
The following human HNSCC cells were used in this study: FaDu (derived from a primary lesion of hypopharyngeal SCC), SAS (derived from a primary lesion of tongue SCC) and HSC3 (derived from a lymph node metastasis of tongue SCC). Cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum in a humidified 5% CO2 atmosphere at 37 °C.
Quantitative real-time reverse transcription PCR
cDNA synthesis and PCR procedures were described in our previous reports.10, 19 The expression levels of miR-1 (assay ID: 002222) and miR-206 (assay ID: 000510), miR-133a (assay ID: 002246) and miR-133b (assay ID: 002247) were analyzed by Taqman quantitative real-time reverse trancription (RT)-PCR (Taqman MicroRNA Assay; Applied Biosystems, Foster City, CA, USA) and normalized to RNU48 (assay ID: 001006). Taqman probes and primers for EGFR (P/N: Hs01076090_m1), c-MET (P/N: Hs01565575_m1) and GUSB (P/N: Hs99999908_ml) as an internal control were obtained from Applied Biosystems.
Mature miRNA transfection
To perform gain-of-function assays, we used the following mature miRNAs species: Pre-miR miRNA precursors (miR-1; P/N: PM10633, miR-206; P/N: PM10409, and negative control miR; P/N: AM17111; Applied Biosystems). The miRNA transfection procedures and confirmation of miRNA transfection efficiency were described in our previous reports.10, 19, 20
Cell proliferation, migration and invasion assays
Cell proliferation was determined with the XTT assay using a Cell Proliferation Kit II (Roche Applied Sciences, Tokyo, Japan). Cell migration activity was analyzed using uncoated Transwell polycarbonate membrane filters. Cell invasion was evaluated using modified Boyden chambers containing Transwell membrane filter inserts precoated with Matrigel. These assays were carried out as previously described.14, 19, 20
Identification of genes putatively regulated by miR-1 and miR-206
To investigate genes putatively regulated by miR-1 and miR-206, we used the TargetScan database (Release 6.2, http://www.targetscan.org/). Next, to identify upregulated genes in HNSCC clinical specimens, we used Gene Expression Omnibus (GEO; accession number: GSE9638). Finally, putative candidate genes were categorized in the Kyoto Encyclopedia of Genes and Genomics (KEGG) using the GeneCodis program. The strategy behind this analysis procedure was described previously.8, 9, 10, 12, 13, 14
Immunohistochemistry
A total of 23 formalin-fixed paraffin-embedded tissues were used. The patients’ backgrounds and clinicopathological characteristics are summarized in Table 2. The tissues were immunostained following the manufacturer’s protocol for the Ultra-Vision Detection system (Thermo Scientific, Fremont, CA, USA). The primary rabbit polyclonal antibodies against EGFR (#4267; Cell Signaling, Danvers, MA, USA) and c-MET (#8198; Cell Signaling) were diluted 1:50 and 1:300, respectively. The slides were treated with biotinylated goat anti-rabbit antibodies. The procedure for immunohistochemistry was described previously.21, 22
Western blotting
Immunoblotting was performed as follows with rabbit antibodies obtained from Cell Signaling: anti-EGFR antibody (1:1000, #4267), anti-c-MET antibody (1:1000, #8198), anti-p-EGFR antibody (1:1000, #2237), anti-p-c-MET antibody (1:1000, #3077), anti-Akt antibody (1:1000, #4691), anti-p-Akt antibody (1:1000, #4060), anti-Erk1/2 antibody (1:1000, #4695) and anti-p-Erk1/2 antibody (1:2000, #4370). Anti-GAPDH antibodies (1:1000, ab8245; Abcam, Cambridge, UK) were used as an internal control. The membrane was washed and incubated with anti-rabbit IgG, HRP-linked antibody (#7074; Cell Signaling). Specific complexes were visualized with the echochemiluminescence detection system (GE Healthcare, Little Chalfont, UK). The procedure for western blotting was described in previous studies.19, 20, 21
Plasmid construction and dual-luciferase reporter assays
The partial wild-type sequences of the EGFR and c-MET 3′-untranslated regions (UTR) or those with a deleted miR-1 and miR-206 target site (positions 746–752 of the EGFR and 499–505 and 814–820 of the c-MET 3′-UTR) were inserted between the Xhol-Pmel restriction sites in the 3′-UTR of the hRluc gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). The procedure for the dual-luciferase assay was described previously.19, 20, 21
Statistical analysis
The relationships between two groups and the numerical values obtained by qPCR were analyzed using the paired t-test. Spearman’s rank test was used to evaluate the correlation between the expression of miR-1, miR-206 and target genes. The relationships among more than three variables and numerical values were analyzed using the Bonferroni-adjusted Mann–Whitney U-test. All analyses were performed using Expert StatView (version 4, SAS Institute Inc., Cary, NC, USA).
Results
Expression of miR-1 and miR-206 in clinical HNSCC specimens and cell lines
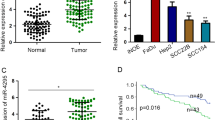
To validate our previously established miRNA expression signature of HNSCC, we evaluated miR-1 and miR-206 expression in 22 clinical HNSCC specimens. The expression levels of miR-1 and miR-206 were significantly reduced in HNSCC cell lines and cancer tissues compared with corresponding adjacent non-cancerous epithelia (Figures 1a and b). In human genome, miR-1/miR-133a and miR-206/miR-133b forms clustered miRNAs in 20q13.33 and 6p12.1, respectively. We also evaluated the expression levels of miR-133a and miR-133b and confirmed downregulation of these miRNAs in HNSCC specimens (Figures 1a and b). Spearman’s rank test showed a positive correlation between the expression of miR-1/miR-133a and miR-206/miR-133b (Figures 1a and b).
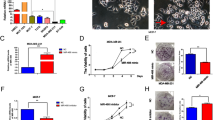
Expression levels of miR-1/miR-133a and miR-206/miR-133b in head and neck squamous cell carcinoma (HNSCC) clinical specimens and functional significance of miR-1 and miR-206 in HNSCC cells. (a) Expression levels of miR-1 and miR-133a in HNSCC clinical specimens and cell lines. RNU48 was used as an internal control. N, noncancerous tissues; T, tumor tissues. Spearman’s rank test showed a positive correlation between the expression of miR-1 and that of miR-133a (r=0.921 and P<0.0001). (b) Expression levels of miR-206 and miR-133b in HNSCC clinical specimens and cell lines. Spearman’s rank test showed a positive correlation between the expression of miR-206 and that of miR-133b (r=0.872 and P<0.0001). (c) Cell proliferation was determined by XTT assay 72 h after transfection with miR-1 or miR-206. (d) Cell movement was assessed by migration assay 48 h after transfection with miR-1 or miR-206. (e) Cell invasion was characterized by invasion assay 48 h after transfection with miR-1 or miR-206. *P<0.0001.
Effect of restoring miR-1 and miR-206 on cell proliferation, migration and invasion activities of HNSCC cell lines
To investigate the functional roles of miR-1 and miR-206, we performed gain-of-function assays using miRNA transfection into three HNSCC cell lines (FaDu, SAS and HSC3).
The XTT assay demonstrated that cell proliferation was significantly inhibited in miR-1 and miR-206 transfectants in comparison with the mock or miR-control transfectant cells (Figure 1c).
The migration assay demonstrated that cell migration activity was significantly inhibited in miR-1 and miR-206 transfectants in comparison with the mock or miR-control transfectant cells (Figure 1d).
The Matrigel invasion assay demonstrated that cell invasion activity was significantly inhibited in miR-1 and miR-206 transfectants in comparison with the mock or miR-control transfectant cells (Figure 1e).
Selection of candidate target genes regulated by miR-1 and miR-206 in HNSCC cells
We performed in silico and gene expression analyses to identify genes targeted by both miR-1 and miR-206 for regulation. Our strategy to narrow down miR-1- and miR-206-regulated genes is shown in Figure 2. First, we selected putative miR-1 and miR-206 target genes using the TargetScan database and identified 4246 genes. The gene set was then analyzed with a publicly available gene expression data set in GEO (accession number: GSE9638) and genes upregulated (log2 ratio >1.0) in HNSCC were chosen. A total of 494 genes were identified as candidate targets of miR-1 and miR-206 regulation.
Next, these genes were then categorized into KEGG pathways using GeneCodis analysis and 13 pathways were identified as significantly enriched (Supplementary Table 1). Among these pathways, we focused on the top five based on the number of genes: pathways in cancer, focal adhesion pathways, MAPK signaling pathways, regulation of actin cytoskeleton pathways and ECM–receptor interaction pathways. The genes involved in these pathways are listed in Supplementary Tables 2–6. A total of 32 genes were involved in these pathways, and we focused on EGFR and c-MET because these two tyrosine kinase receptors are deeply involved in HNSCC pathogenesis.23, 24, 25
The expression status of EGFR and c-MET in HNSCC clinical specimens
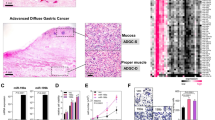
First, we investigated the expression status of EGFR and c-MET proteins in HNSCC clinical specimens using immunohistochemical staining. A total of 23 specimens were checked in this study, and 22 samples stained positively for EGFR (⩾50% of positive cells with moderate or strong staining) and two samples were positive for c-MET. Two samples stained positively for both EGFR and c-MET (Figure 3). Clinicopathological characteristics are summarized in Table 2.
Immunohistochemical staining of epidermal growth factor receptor (EGFR) and c-MET in head and neck squamous cell carcinoma (HNSCC) clinical specimens. EGFR and c-MET expression are different in cancer lesions and adjacent non-cancer tissues in the same fields (original magnification, x40). Patient numbers 1 and 11: EGFR and c-MET stained positively; patient number 2: only EGFR was positive; patient number 23: EGFR and c-MET negative.
We also investigated the mRNA expression levels of EGFR and c-MET in 22 HNSCC clinical specimens by quantitative real-time RT-PCR. EGFR and c-MET were significantly upregulated in HNSCC tumor tissues (Figures 4a and b). Spearman’s rank test showed a negative correlation between the expression of EGFR and miR-1 (P<0.0001, R=−0.0643, Figure 4a), EGFR and miR-206 (P<0.0001, R=−0.657, Figure 4a), c-MET and miR-1 (P=0.0016, R=−0.482, Figure 4b) and c-MET and miR-206 (P=0.0083, R=−0.402, Figure 4b).
Expression levels of epidermal growth factor receptor (EGFR) and c-MET in head and neck squamous cell carcinoma (HNSCC) clinical specimens. (a) Messenger RNA (mRNA) expression levels of EGFR in HNSCC clinical specimens. GUSB was used as an internal control. The negative correlation between EGFR expression and miR-1 or miR-206. Spearman’s rank test was used to evaluate the correlation. (b) mRNA expression levels of c-MET in HNSCC clinical specimens. GUSB was used as an internal control. The negative correlation between c-MET expression and miR-1 or miR-206.
Direct regulation of EGFR and c-MET by miR-1 and miR-206 in HNSCC cells
We investigated whether EGFR and c-MET expression would be reduced by restoration of miR-1 or miR-206 in HNSCC cells. The mRNA and protein levels of expression of EGFR were significantly reduced by miR-1 or miR-206 transfection compared with that in mock- or miR-control-transfected cells (Figures 5a and b). Similar to EGFR expression, expression levels of c-MET were significantly reduced by miR-1 or miR-206 transfection (Figures 6a and b).
Regulation of epidermal growth factor receptor (EGFR) expression by miR-1 and miR-206 in head and neck squamous cell carcinoma (HNSCC) cell lines. (a) EGFR gene expression 72 h after transfection with 10 nm miR-1 or miR-206 into SAS cells. GAPDH was used as an internal control. *P<0.0001. (b) EGFR protein expression 72 h after transfection with miR-1 or miR-206. GAPDH was used as a loading control. (c) Luciferase reporter assay using the vectors encoding putative miR-1 or miR-206 target sites of the EGFR 3′-untranslated region for both wild-type and mutant cotransfectants. Renilla luciferase values were normalized to firefly luciferase values. *P<0.0001.
Regulation of c-MET expression by miR-1 and miR-206 in head and neck squamous cell carcinoma (HNSCC) cell lines. (a) c-MET gene expression 72 h after transfection with 10 nm miR-1 or miR-206. GAPDH was used as an internal control. *P<0.0001. (b) c-MET protein expression 72 h after transfection with 10 nm miR-1 or miR-206. GAPDH was used as the loading control. (c) Luciferase reporter assay using the vectors encoding putative miR-1 or miR-206 target sites of the c-MET 3′-untranslated region for both wild-type and mutant cotransfectants. Renilla luciferase values were normalized to firefly luciferase values. *P<0.0001.
Furthermore, we performed luciferase reporter assay in SAS cells to determine whether EGFR and c-MET mRNA contained target sites for miR-1 and miR-206. We used vectors encoding either a partial wild-type sequence or a sequence in which the miRNA binding site had been mutated from the 3′-UTR of EGFR and c-MET mRNAs. We found that the luminescence intensity was significantly reduced by co-transfection with miR-1, miR-206 and the vector carrying the wild-type 3′-UTR of EGFR and c-MET mRNA (Figures 5c and 6c).
Effects of EGFR and c-MET oncogenic signaling by miR-1 and miR-206 transfection
We analyzed the effects of downstream oncogenic signaling of EGFR and c-MET by using either miR-1 or miR-206 transfectants. The phosphorylation status of EGFR (Tyr1045), c-MET (Tyr1234/1235), AKT (Ser 473) and ERK1/2 (Thr 202/yr 204) were examined. Restoration of miR-1 and miR-206 reduced phosphorylation of EGFR, c-MET, AKT and ERK1/2 in SAS cells (Figure 7).
Effects of miR-1 and miR-206 transfection on epidermal growth factor receptor (EGFR) and c-MET oncogenic signaling. Western blot analysis showed that miR-1 and miR-206 expression led to reductions in phosphorylation of EGFR, c-MET, AKT and Erk1/2 protein in SAS cells. GAPDH was used as a loading control.
Discussion
Our miRNA expression signatures of HNSCC showed that clustered miRNAs, miR-1/miR-133a and miR-206/miR-133b were significantly downregulated in cancer tissues.9, 11 Our present data confirmed downregulation of miR-1 and miR-206 in HNSCC clinical specimens. In the human genome, miR-1-1/miR-133a-2, miR-1-2/miR-133a-1 and miR-206/miR-133b form clustered miRNAs and these clusters are located at three different chromosomal loci, that is, 20q13.33, 18q11.2 and 6p12.1, respectively.18 The mature sequences of miR-1-1 and miR-1-2 are identical, but miR-1 and miR-206 differ by four nucleotides (miRBase: release 21). The seed sequences of miR-1-1/miR-1-2/miR-206 are identical, suggesting that these miRNAs regulate the same target genes in cancer or non-cancerous cells. The molecular mechanisms responsible for the silencing of miR-1 and miR-206 in cancer cells are not clear. Past study indicated that miR-1-1 contains CpG islands (spanning exon 1 and intron 1) and these sites were methylated in hepatocellular carcinoma cell lines and in primary human hepatocellular carcinomas.26 Moreover, suppression of miR-1 expression was recovered by treatment with a DNA hypomethylating agent and histone deacetylase inhibitor in hepatocellular carcinoma and lung cancer cells.26, 27 The expression control of miR-206 is still unclear. Past study showed that transcriptional factor activator protein 1 binding site was existed in the promoter region of miR-206 in human genome and activator protein 1-induced miR-206 expression. In contrast, a transcriptional repressor protein (YY1) was repressed expression of miR-206.28 Further studies are needed to understand the molecular mechanisms of expression control of these miRNAs in cancer cells.
The functional significance of miR-1 and miR-206 in HNSCC cells has been investigated by ectopic expression of these miRNAs in cancer cells. Our data showed that ectopic expression of miR-1 and miR-206 significantly suppressed cancer cell aggressiveness, indicating that these miRNAs act as tumor suppressors in normal cells. The tumor-suppressive role of miR-1 was reported in several types of cancers, such as lung cancer, colon cancer, genitourinary cancer, head and neck cancer and sarcoma.20, 29, 30, 31, 32 Downregulation of miR-1 has been observed in multiple cancers, suggesting that cancer pathways regulated by miR-1 or their targets could provide new insights into potential molecular mechanisms of cancer pathogenesis. Like miR-1, the downregulation of miR-206 and its anti-tumor roles were reported for several cancers.18, 21, 31, 33 Ectopic miR-206 expression induced cell cycle arrest and apoptosis in breast cancer, prostate cancer, lung cancer, gastric cancer, sarcoma, glioma and neuroblastoma.18, 31, 34 Expression levels of miR-206 were correlated with an advanced stage, lymph node metastasis and poor prognosis in several cancers.34, 35 In oral SCC cells, K-Ras was directly regulated by miR-206.33 Activation of the K-Ras oncogene is implicated in several types of cancers36, 37, suggesting that miR-206 acts as a critical anti-tumor effector in oral SCC cells.
We hypothesized that identification of the molecular targets and pathways regulated by tumor-suppressive miR-1 and miR-206 in HNSCC would enhance our understanding of the disease and could suggest more effective strategies for future therapeutic interventions. Thus, we identified the genetic targets regulated by miR-1 and miR-206. In this screening, several pathways and targets were annotated as subjects of miR-1 and miR-206 regulation in HNSCC cells. Among them, we focused on two receptor tyrosine kinase (RTK) genes (EGFR and c-MET) because these receptors mediate signals contributing to cancer cell aggressiveness and drug resistance.21, 25, 27, 30 Our data showed that EGFR and c-MET were directly regulated by miR-1 and miR-206, making them tumor-suppressive miRNAs. Moreover, ectopic expression of these miRNAs reduced the levels of p-ERK1/2 and p-ATK as downstream signaling molecules of these RTKs. Previous studies demonstrated the inhibition of c-MET by miR-1 in several cancers.30, 31, 38 More recently, we showed that ectopic expression of miR-206 suppressed dual tyrosine kinase receptors, EGFR and c-MET in lung squamous cell carcinoma cells.21 Past studies showed that EGFR was regulated by miR-133a and miR-133b as clustered miRNAs in miR-1 and miR-206, respectively.39, 40 However, we had no positive data expression control of EGFR by miR-133a or miR-133b in HNSCC cells (data not shown).
Overexpression of EGFR is observed in ~90% of patients with HNSCC and aberrant activation of EGFR signaling enhances proliferation, invasion, metastasis and angiogenesis of cancer cells.23 Aberrant expression of EGFR correlates with decreased survival. Furthermore, EGFR and its mediated signal cascade have been implicated in the pathogenesis of HNSCC.24 In 2006, the EGFR-targeting monoclonal antibody cetuximab was approved by Food and Drug Administration (FDA) for treatment of patients with HNSCC. Currently, cetuximab is approved in combination with radiation therapy for HNSCC as a first-line treatment, or with other anticancer drugs in recurrent or metastatic disease.41, 42 However, the curative effects of cetuximab treatments are limited, and it is difficult to achieve complete remission in this disease. Many studies have suggested that several alternative signal cascades are activated by EGFR blockade therapies in cancer cells.43, 44 Interestingly, several reports showed that cetuximab induced c-MET signal activation, suggesting a possible mechanism of acquired resistance to EGFR inhibitors.45 Recent studies indicate that c-MET activation is responsible for ~20% of resistance to EGFR inhibitors.46
Similar to EGFR expression, aberrant expression of c-MET was observed in 90% of HNSCC cell lines and 84% of HNSCC patient samples.25 Our present data showed that overexpression of both EGFR and c-MET occurred in several HNSCC clinical specimens. It is extremely important to realize that both tumor-suppressive miR-1 and miR-206 inhibited EGFR and c-MET oncogenic signaling in HNSCC cells. Therefore, we propose that therapeutic strategies relying on dual blocking of EGFR and c-MET oncogenic signaling are indispensable for HNSCC treatment. The identification of novel molecular pathways regulated by miR-1 and miR-206 may lead to a better understanding of HNSCC aggressiveness and mechanisms of drug resistance.
In conclusion, downregulation of miR-1 and miR-206 enhances cancer cell aggressiveness through their targeting of tyrosine kinase receptors, EGFR and c-MET in HNSCC cells. Ectopic expression of miR-1 and miR-206 inhibited EGFR and c-MET downstream signal cascades. Elucidation of the cancer pathways and target genes regulated by tumor-suppressive miR-1 and miR-206 should provide new approaches and potential therapeutic targets in the treatment of HNSCC.
Accession codes
References
Leemans, C. R., Braakhuis, B. J. & Brakenhoff, R. H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 11, 9–22 (2011).
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 (2008).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 (2006).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Wiemer, E. A. The role of microRNAs in cancer: no small matter. Eur. J. Cancer 43, 1529–1544 (2007).
Iorio, M. V. & Croce, C. M. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 27, 5848–5856 (2009).
Fukumoto, I., Hanazawa, T., Kinoshita, T., Kikkawa, N., Koshizuka, K., Goto, Y. et al. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer 112, 891–900 (2015).
Fukumoto, I., Kinoshita, T., Hanazawa, T., Kikkawa, N., Chiyomaru, T., Enokida, H. et al. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br. J. Cancer 111, 386–394 (2014).
Kikkawa, N., Hanazawa, T., Fujimura, L., Nohata, N., Suzuki, H., Chazono, H. et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). Br. J. Cancer 103, 877–884 (2010).
Nohata, N., Hanazawa, T., Kikkawa, N., Sakurai, D., Fujimura, L., Chiyomaru, T. et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br. J. Cancer 105, 833–841 (2011).
Fukumoto, I., Kikkawa, N., Matsushita, R., Kato, M., Kurozumi, A., Nishikawa, R. et al. Tumor-suppressive microRNAs (miR-26a/b, miR-29a/b/c and miR-218) concertedly suppressed metastasis-promoting LOXL2 in head and neck squamous cell carcinoma. J. Hum. Genet. 61, 109–118 (2016).
Kinoshita, T., Hanazawa, T., Nohata, N., Kikkawa, N., Enokida, H., Yoshino, H. et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget 3, 1386–1400 (2012).
Kinoshita, T., Nohata, N., Hanazawa, T., Kikkawa, N., Yamamoto, N., Yoshino, H. et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br. J. Cancer 109, 2636–2645 (2013).
Gilcrease, M. Z. Integrin signaling in epithelial cells. Cancer Lett. 247, 1–25 (2007).
Kim, S. H., Turnbull, J. & Guimond, S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 209, 139–151 (2011).
Kurozumi, A., Goto, Y., Matsushita, R., Fukumoto, I., Kato, M., Nishikawa, R. et al. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 107, 84–94 (2015).
Nohata, N., Hanazawa, T., Enokida, H. & Seki, N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget 3, 9–21 (2012).
Nohata, N., Hanazawa, T., Kikkawa, N., Mutallip, M., Sakurai, D., Fujimura, L. et al. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J. Hum. Genet. 56, 595–601 (2011).
Nohata, N., Sone, Y., Hanazawa, T., Fuse, M., Kikkawa, N., Yoshino, H. et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget 2, 29–42 (2011).
Mataki, H., Seki, N., Chiyomaru, T., Enokida, H., Goto, Y., Kumamoto, T. et al. Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. Int. J. Oncol. 46, 1039–1050 (2015).
Koeppen, H., Yu, W., Zha, J., Pandita, A., Penuel, E., Rangell, L. et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib+/-onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin. Cancer Res. 20, 4488–4498 (2014).
Kalyankrishna, S. & Grandis, J. R. Epidermal growth factor receptor biology in head and neck cancer. J. Clin. Oncol. 24, 2666–2672 (2006).
Chung, C. H., Ely, K., McGavran, L., Varella-Garcia, M., Parker, J., Parker, N. et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J. Clin. Oncol. 24, 4170–4176 (2006).
Seiwert, T. Y., Jagadeeswaran, R., Faoro, L., Janamanchi, V., Nallasura, V., El Dinali, M. et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 69, 3021–3031 (2009).
Datta, J., Kutay, H., Nasser, M. W., Nuovo, G. J., Wang, B., Majumder, S. et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 68, 5049–5058 (2008).
Nasser, M. W., Datta, J., Nuovo, G., Kutay, H., Motiwala, T., Majumder, S. et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 283, 33394–33405 (2008).
Song, G. & Wang, L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS ONE 4, e6880 (2009).
Mataki, H., Enokida, H., Chiyomaru, T., Mizuno, K., Matsushita, R., Goto, Y. et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J. Hum. Genet. 60, 53–61 (2015).
Migliore, C., Martin, V., Leoni, V. P., Restivo, A., Atzori, L., Petrelli, A. et al. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin. Cancer Res. 18, 737–747 (2012).
Yan, D., Dong Xda, E., Chen, X., Wang, L., Lu, C., Wang, J. et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J. Biol. Chem. 284, 29596–29604 (2009).
Yoshino, H., Chiyomaru, T., Enokida, H., Kawakami, K., Tatarano, S., Nishiyama, K. et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br. J. Cancer 104, 808–818 (2011).
Lin, F., Yao, L., Xiao, J., Liu, D. & Ni, Z. MiR-206 functions as a tumor suppressor and directly targets K-Ras in human oral squamous cell carcinoma. Onco Targets Ther. 7, 1583–1591 (2014).
Zhang, L., Liu, X., Jin, H., Guo, X., Xia, L., Chen, Z. et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 332, 94–101 (2013).
Yang, Q., Zhang, C., Huang, B., Li, H., Zhang, R., Huang, Y. et al. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur. J. Gastroenterol. Hepatol. 25, 953–957 (2013).
Rajalingam, K., Schreck, R., Rapp, U. R. & Albert, S. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta 1773, 1177–1195 (2007).
Caulin, C., Nguyen, T., Longley, M. A., Zhou, Z., Wang, X. J. & Roop, D. R. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. 64, 5054–5058 (2004).
Han, C., Zhou, Y., An, Q., Li, F., Li, D., Zhang, X. et al. MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol. 36, 6715–6723 (2015).
Liu, L., Shao, X., Gao, W., Zhang, Z., Liu, P., Wang, R. et al. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 279, 3800–3812 (2012).
Cui, W., Zhang, S., Shan, C., Zhou, L. & Zhou, Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 280, 3962–3974 (2013).
Bonner, J. A., Harari, P. M., Giralt, J., Azarnia, N., Shin, D. M., Cohen, R. B. et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 354, 567–578 (2006).
Vermorken, J. B., Mesia, R., Rivera, F., Remenar, E., Kawecki, A., Rottey, S. et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 359, 1116–1127 (2008).
Bertotti, A. & Sassi, F. Molecular pathways: sensitivity and resistance to anti-EGFR antibodies. Clin. Cancer Res. 21, 3377–3383 (2015).
Rabinowits, G. & Haddad, R. I. Overcoming resistance to EGFR inhibitor in head and neck cancer: a review of the literature. Oral Oncol. 48, 1085–1089 (2012).
Madoz-Gurpide, J., Zazo, S., Chamizo, C., Casado, V., Carames, C., Gavin, E. et al. Activation of MET pathway predicts poor outcome to cetuximab in patients with recurrent or metastatic head and neck cancer. J. Transl. Med. 13, 282 (2015).
Ma, P. C., Tretiakova, M. S., MacKinnon, A. C., Ramnath, N., Johnson, C., Dietrich, S. et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 47, 1025–1037 (2008).
Acknowledgements
This study was supported by the KAKENHI (C), 15K10800, 15K10801, 25462676 and 26462596.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Koshizuka, K., Hanazawa, T., Fukumoto, I. et al. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet 62, 113–121 (2017). https://doi.org/10.1038/jhg.2016.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.47
This article is cited by
-
Non-coding RNAs as potential therapeutic targets for receptor tyrosine kinase signaling in solid tumors: current status and future directions
Cancer Cell International (2024)
-
Study of MicroRNA (miR-221-3p, miR-133a-3p, and miR-9-5p) Expressions in Oral Submucous Fibrosis and Squamous Cell Carcinoma
Indian Journal of Clinical Biochemistry (2023)
-
Emerging roles and mechanisms of miR-206 in human disorders: a comprehensive review
Cancer Cell International (2022)
-
The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer
Cancer Cell International (2019)
-
MiR-206 inhibits epilepsy and seizure-induced brain injury by targeting CCL2
Cytotechnology (2019)