Abstract
Y chromosome microsatellite (Y-STR) diversity has been studied in different Mongolic-speaking populations from South Siberia, Mongolia, North-East China and East Europe. The results obtained indicate that the Mongolic-speaking populations clustered into two groups, with one group including populations from eastern part of South Siberia and Central Asia (the Buryats, Barghuts and Khamnigans) and the other group including populations from western part of Central Asia and East Europe (the Mongols and Kalmyks). High frequency of haplogroup C3-M407 (>50%) is present in the Buryats, Barghuts and Khamnigans, whereas in the Mongols and Kalmyks its frequency is much lower. In addition, two allelic combinations in DYS385a,b loci of C3-M407 haplotypes have been observed: the combination 11,18 (as well as 11,17 and 11,19) is frequent in different Mongolic-speaking populations, but the 11,11 branch is present mainly in the Kalmyks and Mongols. Results of locus-specific sequencing suggest that the action of gene conversion is a more likely explanation for origin of homoallelic 11,11 combination. Moreover, analysis of median networks of Y-STR haplotypes demonstrates that at least two gene conversion events can be revealed—one of them has probably occurred among the Mongols, and the other event occurred in the Barghuts. These two events give an average gene conversion rate range of 0.24–7.1 × 10–3 per generation.
Similar content being viewed by others
Introduction
Modern Mongolic-speaking populations have a wide geographic dispersal, encompassing northern parts of Central and Eastern Asia, South Siberia and East Europe. Analysis of genome-wide genotype data has shown that South Siberia and Mongolia area appears to be a homeland for Turkic and related Mongolic ancestors, from where the historically recorded Turkic nomadic migrations and later Mongol expansion were carried out during the period from ninth to seventeenth centuries.1 Results of analysis of maternal (mitochondrial DNA) and paternal (Y chromosome) gene pools demonstrate a close relationship between the Mongolic-speaking populations of South Siberia, Mongolia, North-East China and East Europe, thus suggesting their origin from a common ancestral gene pool.2, 3, 4, 5 Among the Y chromosome lineages, a relatively high frequency of haplogroup C3-M407 has been revealed in all Mongolic-speaking populations studied to date.2, 3, 6 However, remarkable between-population differences were found for distribution of C3-M407 haplotypes carrying mutations at the duplicated tetranucleotide microsatellite DYS385a,b lying in palindrome P4.2, 3, 6
It is known that the male-specific region of the human Y chromosome contains eight large inverted repeats (palindromes), each with two repeat copies (arms) separated by single-copy spacers.7 These duplicated segments differ only slightly (<0.1% differences) because of frequent gene conversion events homogenizing variants that arise through the slow processes of nucleotide substitution and insertion/deletion at the two loci.8, 9, 10 Highly polymorphic DNA markers (such as microsatellite or short tandem repeat (STR) loci) should be useful for studying of gene conversion in Y chromosome. One of the most variable Y chromosome STR systems (Y-STRs) is DYS385a,b loci, which exist in two polymorphic copies lying in palindrome P4. It is known that the inverted repeat containing this system represents the duplicated ~190 kb repeats, separated by ~40 kb of unique spacer sequence.7 In general, the distal copy is known as DYS385a and the proximal copy as DYS385b.
Recently, multiple gene conversion events have been revealed in two Y chromosome haplogroups carrying DYS385a,b homoallelic combinations equally with heteroallelic ones–haplogroups O3e and R2.10 For example, haplogroup O3e has a modal combination of 13,18, but also contains both 13,13 and 18,18 combinations. That study has shown that appearance of homoallelic combinations in Y chromosome haplogroups was due to the action of gene conversion. Earlier, it has been found that haplogroup C3-M407 Y chromosomes characteristic mainly of the Mongolic-speaking populations carry not only the combinations 11,17, 11,18 and 11,19, but also the combination 11,11.6 The overwhelming majority of C3-M407 haplotypes in populations of South Siberia and Central Asia are defined by allelic combination 11,18 (or sometimes 11,17 and 11,19), but the 11,11 branch is present in the Kalmyks, Mongols, Buryats, Tuvinians and Altaians.2, 3, 6 Because a transition from 11,18 to 11,11 via multiple single-step mutation events on otherwise similar haplotypic backgrounds seems unlikely, gene conversion is a more appropriate explanation (as suggested by Balaresque et al.10), although such causes as deletion of DYS385a or DYS385b locus or mutation in a primer-binding site cannot be ruled out.
In order to gain more insights into this problem, we explored haplogroup C3-M407 Y chromosomes in the Mongolic-speaking populations of the Barghuts, Buryats and Kalmyks, using 29 Y-STRs and single-nucleotide polymorphism typing of Y chromosome haplogroup-specific sites. In addition, the genetic relationships between the Mongolic-speaking populations from South Siberia, Mongolia, North-East China and East Europe were examined based on Y chromosome STR diversity.
Materials and methods
Subjects and DNA typing
Blood samples from 76 unrelated Barghut males were collected in different localities of Hulun Buir Aimak, Inner Mongolia, China. Total DNA was extracted by the standard phenol/chloroform method. All the samples were collected after obtaining written informed consent, with ethical approval at the Institute of Biological Problems of the North FEB RAS in Magadan, Russia.
Barghut samples were typed for 17 Y-STR loci (DYS19, DYS385a, DYS385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, GATA-H4, DYS448, DYS456, DYS458, DYS635) using the AmpFl-STR YFiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Products of amplification were analyzed on ABI 3500xL Genetic Analyzer (Applied Biosystems). Electrophoresis results were analyzed using the GeneMapper Software v. 4.1 (Applied Biosystems).
The Y chromosome binary markers were typed in the Barghuts as described elsewhere2 through direct sequencing on ABI 3130 and ABI 3500xL Genetic Analyzers (Applied Biosystems) or by analysis of restriction fragment-length polymorphisms using PCR primers summarized in Karafet et al.11 Refined nomenclature of the Y chromosome variants suggested by Karmin et al.12 has been used for haplogroup designations.
Haplogroup C3-M407 samples from populations of the Buryats previously typed for 12 Y-STR loci (DYS19, DYS385a, DYS385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439) using PowerPlex Y System (Promega, Madison, WI, USA)13 were re-typed for 17 Y-STR loci. Haplogroup C3-M407 samples from populations of the Barghuts, Buryats and Kalmyks were typed for 13 rapidly mutating Y-STRs (RM Y-STRs), which were amplified in a single multiplex reaction as described by Rogalla et al.14 This assay encompasses 13 markers (DYS449, DYS458, DYS516, DYS518, DYS526b, DYS534, DYS547, DYS570, DYS576, DYS611, DYS612, DYS626 and DYS627).
Locus-specific amplification of DYS385 was carried out using forward primers DYS385a and DYS385b suggested by Kittler et al.15 and reverse DYS385-R primer reported by Park et al.16 These primers generate a single product of about 800 bp in length for each STR locus. DNA sequencing of various length alleles of both loci was performed on ABI 3500xL Genetic Analyzer using Big Dye chemistry (Applied Biosystems) according to the recommendations of the manufacturer. Different DYS385a,b allelic combinations—11,11, 11,12, 11,17, 11,18 and 12,19—were sequenced.
Statistical analysis
The allele sizes for DYS389II were determined with the subtraction of DYS389I, and both loci were included in the calculations. The Barghut population data set was compared with previously described populations of Siberia, Central and Eastern Asia belonging to different groups of Altaic family of languages, for which data for 12 Y-STR loci (DYS19, DYS385a, DYS385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439) were available. These populations included Mongolic-speaking Buryats,13 Kalmyks, Mongols and Khamnigans,2 Turkic-speaking Altaian Kazakhs,17 Sojots, Southern Altaians, Khakassians and Tuvinians2 and Tungusic-speaking Evenks and Manchus.2, 18 Tibetan-speaking Tibetans have also been used for population comparisons.19 In addition, four populations (Turkic-speaking Teleuts, Tofalars and Shors and Tungusic-speaking Evens) typed for the above 12 Y-STR loci using PowerPlex Y System (Promega) were also analyzed. These population samples have been partially published by us,6, 20, 21, 22 but the complete set of Y-STR profiles is included here in Supplementary Table S1.
Summary statistics were calculated using ARLEQUIN 3.5.23 Genetic differentiation between populations was estimated by means of distance methods based on the number of different alleles of the microsatellite (FST) and the sum of the squared number of repeat differences between two alleles (RST). The measure RST differs from FST in taking into account of the mutation process at microsatellite loci, for which a generalized stepwise mutation model appears appropriate.24 The software ARLEQUIN 3.5 was used to perform Analysis of Molecular Variation (AMOVA). The statistical significance tests for pairwise FST and RST values were performed at 1000 permutations and for AMOVA at 10000 permutations. Loci DYS385a and DYS385b were excluded from analysis of between-population relationships, because an unambiguous assignment of the alleles to these loci is impossible without their separate typing. In addition, we excluded DYS19 because it is duplicated in some haplogroups, particularly in haplogroup C3-M77.
Between-population distances (in the form of FST or RST values) were illustrated by creating a multidimensional scaling plot using the software package STATISTICA v. 7.1 (StatSoft, Inc., Tulsa, OK, USA). Because some Mongolic-speaking populations are only available from the YHRD database,25 we have used an AMOVA tool of the YHRD (release 49) to measure the genetic distances between populations (FST and RST statistics) based on 12 Y-STR loci.
The age of STR variation within haplogroups was estimated as the average squared difference in the number of repeats between all current chromosomes and the founder haplotype (formed by the median values of the repeat scores at each STR locus within the haplogroup), averaged over STR loci and divided by means of a mutation rate.26 For 12 Y-STR and 17 Y-STR loci sets, average mutation rates equal to 2.83 × 10−3 and 3.42 × 10−3 per locus per generation were used, respectively. Values of mutation rates were selected from the study by Ballantyne et al.27 We assumed a generation time of 30 years.28, 29, 30 The range of gene conversion rate (in the number of conversion events per generation) was estimated as described in Balaresque et al.10
Median network analysis
Median-joining networks of Y-STR haplotypes belonging to the C3-M407 haplogroup were constructed using the Network 4.6 program (http://fluxus-engineering.com). For the network construction, the weight of each microsatellite variant (character) within the network was assigned to be inversely proportional to the number of mutations at character.31 The weights varied from the default value of 10 at characters with 1 mutation per character to the value of 1 at fast-mutating characters (Supplementary Table S2).
Results
In total, eight Y chromosome haplogroups were identified in the Barghuts (Table 1). However, more than half (55.3%) of the Barghut Y chromosomes belonged to haplogroup C3-M407. N3-Tat and C3*-M217 encompassed 27.6% and 10.5%, respectively. The remaining haplogroups revealed in the Barghuts (G-M201, J2a-M410, T1-M70, O3-M122 and R2a-M124) were singletons.
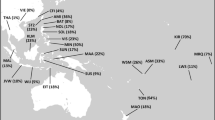
Analysis of 17 Y-STRs provided additional details on haplogroups present in the Barghuts (Supplementary Table S3). To compare the Barghut data with published data sets, we reduced the 17-STR profile to a 9-STR profile (loci DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439) and estimated population pairwise FST and RST distances (Supplementary Table S4). The results of the multidimensional scaling of the FST values presented in Figure 1 illustrate the close genetic affinities between the Mongolic-speaking Barghuts, Buryats and Khamnigans, while the Mongolic-speaking Mongols and Kalmyks as well as the Turkic-speaking Altaian Kazakhs were separated from the above populations. Somewhat different picture emerged from the results of the RST value analysis (Figure 2) demonstrating that populations are grouped into two clusters, with most Mongolic-speaking populations and genetically close to them populations of Altaian Kazakhs, Sojots, Tibetans and Evens being separated from the other populations, such as the Turkic-speaking Altaians, Teleuts, Shors, Khakassians, Tuvinians, Tofalars and Tungusic-speaking Evenks and Manchu. We should note, however, that in both cases Mongolic-speaking populations form two distinguishable groups—eastern group joining the Buryats, Barghuts and Khamnigans, and western group represented by the Mongols and Kalmyks.
Multidimensional scaling (MDS) plot of pairwise FST genetic distances using the Y chromosome STR haplotypes data for populations of Siberia, Central and Eastern Asia. Acronyms: AKZ—Altaian Kazakhs, ALT—Altaians, BAR—Barghuts, BUR—Buryats, EVK—Evenks, EVN—Evens, KHA—Khakassians, KHM—Khamnigans, KM—Kalmyks, MAN—Manchu, MON—Mongols, SHO—Shors, SOJ—Sojots, TEL—Teleuts, TIB—Tibetans, TOF—Tofalars, TUV—Tuvinians. The stress value for the MDS plots is 0.000196.
MDS plot of pairwise RST genetic distances using the Y chromosome STR haplotype data for populations of Siberia, Central and Eastern Asia. Acronyms are as in the Figure 1 legend. The stress value for the MDS plots is 0.000048.
Results of FST-based analysis of 12 Y-STR loci haplotypes from Mongolic-speaking populations available from the YHRD database also indicate that the Buryats and Barghuts differ significantly from other Mongolic-speaking populations of Mongolia and China (Supplementary Figure S1). Meanwhile, RST-based analysis demonstrates that the Barghuts share a smaller distances with the Mongols inhabiting the North-East China (Inner Mongolia and Liaoning) (Supplementary Figure S2).
The AMOVA performed using the 17 populations of South Siberia, Central and Eastern Asia showed that a 16.4% and 19.9% of genetic differentiation (for FST and RST distances, respectively) are due to differences among ethnic groups. Similar FST values were obtained for Mongolic- and Turkic-speaking populations (~15% in both cases), but average RST values were higher in Turkic-speaking populations (18.2% in Turkic-speaking and 10.6% in Mongolic-speaking populations).
As haplogroup C3-M407, most frequent in Mongolic-speaking populations,2, 6, 7 is represented by two types of Y-STR profiles characterizing by DYS385a,b allelic combinations 11,18 and 11,11, we have defined the number of repeats at each of the two duplicated DYS385 loci using locus-specific amplification. As a result, we have found that the distal DYS385a copy has 11 repeats and the proximal DYS385b copy has 18 repeats. In all separate analyses of the 11,11 allelic combinations, both DYS385a and DYS385b loci yielded identical 11-repeat sequences, confirming that single-band 11,11 patterns for DYS385 in haplogroup C3-M407 reflect an underlying 11,11 genotypes, rather than a deletion (that is, 11,null). Therefore, the action of gene conversion at the duplicated DYS385a,b loci is a more likely explanation for origin of homoallelic 11,11 combination.
We performed median network analysis of 17 Y-STR loci (DYS19, DYS385a, DYS385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, GATA-H4, DYS448, DYS456, DYS458 and DYS635) haplotypes belonging to haplogroup C3-M407. Figure 3 illustrates that this haplogroup is differentiated into two subgroups characterized by DYS385a,b combination 11,18 (and sometimes 11,17 and 11,19) and the combination 11,11. Allelic combination 11,11 is present mainly in the Kalmyks and rarely in the Mongols, Tuvinians, Altaians-Teleuts and Barghuts (Table 2). Therefore, most frequent heteroallelic combination DYS385a,b=11,18 should generate homoallelic combination 11,11 in a single conversion event. However, median network analysis demonstrates that the 11,11 combinations have arisen at least twice and, most probably, on the basis of the 11,17 combinations (Figure 3). Most frequent combination 11,11 found in the Kalmyks, Mongols, Tuvinians and Teleuts has been generated on the basis of haplotype carrying the variants 12 at DYS439 and 18 at DYS458 (haplotype Ht1-DYS385b-17), but the 11,11 combination revealed in the Barghuts could have arisen via separate event because its ancestral haplotype (Ht2-DYS385b-17) is characterized by the variants 13 at DYS439 and 17 at DYS458 (Table 3).
A median-joining network based on 17 Y-STR loci haplotypes within the C3-M407 haplogroup. Each circle represents a haplotype, defined by a combination of STR markers. Circle size is shown proportional to haplotype frequency, and the smallest circle represents one haplotype. The lines between circles represent mutational distance, the shortest distance being a single mutational step. Weighting scheme used for MJ network construction is in Supplementary Table S2.
Limited to 17 Y-STRs, median network analysis demonstrates that probable ancestral haplotype displaying the DYS385a,b 11,11 combination in four Kalmyks should originate from the 11,17 haplotype uncovered in four Barghut and Buryat individuals. Correspondingly, independent 11,11 combination found in the Barghuts is most closely related to haplotype carrying the 11,17 combination in two Barghut samples (Table 3). Meanwhile, after combining 17 Y-STR markers with 13 RM Y-STRs only two identical Y-STR profiles were found among the Barghuts carrying the 11,17 combination (samples 17_Bt and 51_Bt), and only two identical profiles were observed in the Kalmyks (Km312 and Km319) displaying the 11,11 combination, although all these individuals were paternally unrelated. However, median network analysis of combined 17 Y-STR and RM Y-STR data sets (Supplementary Table S5) does not allow us to clarify the origin of the 11,11 combination because the connection between the two DYS385a,b combinations has been realized only through the median vectors (Supplementary Figure S3). This is probably due to very high mutation rates of the RM Y-STRs, allowing differentiation of many of the samples at the individual level.
In general, our analysis demonstrates that gene conversion between palindrome arms as well as stepwise mutation processes have an important role in mutational dynamics of the palindromic microsatellite DYS385. Figure 4 shows the most parsimonious scenario represented by combination of stepwise mutational events and gene conversion for haplogroup C3-M407. As expected, the smaller DYS385a locus (with 11 and 12 repeats) has lesser repeat variance than the larger DYS385b locus (with 17, 18 and 19 repeats)—0.0049 versus 0.1055, respectively. This is due to suggested length-dependency of microsatellite mutation rates.32, 33 It is noteworthy, however, that even after gene conversion event (that is, among the 11,11 combinations) DYS385b locus started to expand faster than DYS385a (with the repeat variances 0.01852 versus 0, respectively).
The age of Y-STR variation of haplogroup C3-M407 (having excluded locus DYS385) is about 600 years, while the age of haplotypes carrying the 11,11 combination (~350 years) is slightly smaller than that for the 11,18 combination (~500 years) (Supplementary Table S5). Taking into account two observed gene conversion events for this haplogroup, we have found that an average gene conversion rate range is 0.24–7.1 × 10–3 per generation (Supplementary Table S6).
Discussion
In the present study, we have analyzed Y-STR diversity in different ethnolinguistic groups, including several Mongolic-speaking groups from South Siberia, Mongolia, North-East China and East Europe. The results of the analyses indicate that the Mongolic-speaking populations are differentiated into two groups. The Mongolic-speaking Buryats, Barghuts and Khamnigans cluster together but not with the Mongols and Kalmyks. We should note that similar trend has recently been revealed in studies of genome-wide polymorphisms. Despite the fact that the Mongolic- and Turkic-speaking populations from South Siberia and Mongolia bear an unusually high number of common long chromosomal tracts,1 the Mongolic-speaking Buryats show closer affinities to the Turkic-speaking Altaians and Tuvinians than to the Mongolic-speaking Mongols.34
Ethnologists suggested that the Khamnigans are actually Evenks of Tungusic origin, who were Mongolized in the early sixteenth century, and that the Barghuts have descended from the ancient Turkic-Mongolic tribes inhabiting the Trans-Baikal region during the period from seventh to tenth centuries.35 It was also argued that the Buryats are the descendants of indigenous populations from Lake Baikal who shifted to the Mongolic language.36 For instance, from the sixth to tenth century a large Turkic-speaking tribe, the Kurikans (or Quriqan) were situated around Lake Baikal.37 Later, under the pressure of the Mongol expansion, one part of the Kurikans is assumed to have migrated to the north and the other mixed with Mongolian tribes and stayed in the Trans-Baikal region.36, 37 Meanwhile, results of analysis of maternal and paternal DNA lineages strongly indicate that the Buryats, Barghuts and Khamnigans are closely related genetically.2, 4 Moreover, it has been demonstrated that all Mongolic-speaking peoples of South Siberia, Mongolia, North-East China and East Europe originate from a common ancestral gene pool.2, 3, 4, 5
One of the features of male gene pool of the Mongolic-speaking populations is haplogroup C3-M407. This haplogroup is characteristic of all Mongolic-speaking populations–it is very frequent in the Buryats, Barghuts, Khamnigans and in the Turkic-speaking Sojots (>50%) but not in the Mongols (15.2%) and Kalmyks (10.8%) (Supplementary Table S7). In addition, C3-M407 haplotypes are represented by two allelic combinations in DYS385a,b loci. The combination 11,18 (as well as 11,17 and 11,19) is frequent in different Mongolic-speaking populations, but the 11,11 branch is present mainly in the Kalmyks and Mongols.2, 3, 6 In the present study, we have found that gene conversion is more likely an explanation for origin of homoallelic combination. Moreover, analysis of median networks of Y-STR haplotypes demonstrates that at least two gene conversion events can be revealed—one of them has probably occurred among the West Mongolic ancestors of the Kalmyks, and the other in the Barghuts.
Another scenario suggesting a single gene conversion event with associated parallel mutations at DYS439 and DYS458 occurred in the Barghuts is also possible due to relatively high mutation rates at these loci.29 However, we cannot prefer this explanation because the allelic combination 11,11 is present only in populations of the west of Central Asia and South Siberia, being found among the Mongols, Kalmyks, Buryats from western part of Buryatia, Tuvinians and Altaians-Teleuts.2, 3, 6 This combination is absent among Eastern Buryats and Khamnigans,3, 6 but at a single instance, it has been revealed here in the Barghuts from North-East China, far away from the main area of the 11,11 distribution. Therefore, this fact, along with the results of the median network analysis, allowed us to prefer a model with two gene conversion events that occurred during the expansion of haplogroup C3-M407 in Mongolic-speaking populations. These two events give an average gene conversion rate range of 0.24–7.1 × 10–3 per generation. Our estimate is similar to gene conversion rates for haplogroups O3e and R2 for an upper limit but exceeds significantly a lower limit of ~10−5 events per generation calculated for the above haplogroups.10 This is probably due to much lower age of haplogroup C3-M407. But, in any case our estimate of a lower limit of gene conversion rate is by a factor of 10 slower than single-step mutation rates, in agreement with previous estimates.10 Finally, our research demonstrates that gene conversion should be taken into account in Y chromosome human population studies, being the important force acting on microsatellite evolution in palindromes.
References
Yunusbayev, B., Metspalu, M., Metspalu, E., Valeev, A., Litvinov, S., Valiev, R. et al. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 11, e1005068 (2015).
Malyarchuk, B., Derenko, M., Denisova, G., Khoit, S., Wozniak, M., Grzybowski, T. et al. Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels. J. Hum. Genet. 58, 804–811 (2013).
Kharkov, V. N., Khamina, K. V., Medvedeva, O. F., Simonova, K. V., Eremina, E. R. & Stepanov, V. A. Gene pool of Buryats: clinal variability and territorial subdivision based on data of Y chromosome markers. Russ. J. Genet. 50, 180–190 (2014).
Derenko, M., Malyarchuk, B., Denisova, G., Perkova, M., Rogalla, U., Grzybowski, T. et al. Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of northern Asia and eastern Europe. PLoS ONE 7, e32179 (2012).
Derenko, M., Malyarchuk, B., Grzybowski, T., Denisova, G., Dambueva, I., Perkova, M. et al. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am. J. Hum. Genet. 81, 1025–1041 (2007).
Malyarchuk, B., Derenko, M., Denisova, G., Woźniak, M., Grzybowski, T., Dambueva, I. et al. Phylogeography of the Y-chromosome haplogroup C in northern Eurasia. Ann. Hum. Genet. 74, 539–546 (2010).
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Rozen, S., Skaletsky, H., Marszalek, J. D., Minx, P. J., Cordum, H. S., Waterston, R. H. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003).
Hallast, P., Balaresque, P., Bowden, G. R., Ballereau, S. & Jobling, M. A. Recombination dynamics of a human Y-chromosomal palindrome: Rapid GC-biased gene conversion, multi-kilobase conversion tracts, and rare inversions. PLoS Genet. 9, e1003666 (2013).
Balaresque, P., King, T. E., Parkin, E. J., Heyer, E., Carvalho-Silva, D., Kraaijenbrink, T. et al. Gene conversion violates the stepwise mutation model for microsatellites in Y-chromosomal palindromic repeats. Hum. Mutat. 35, 609–617 (2014).
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L. & Hammer, M. F. New binary polymorphisms reshape and increase resolution of the human Y-chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Karmin, M., Saag, L., Vicente, M., Wilson Sayres, M. A., Järve, M., Talas, U. G. et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 25, 459–466 (2015).
Woźniak, M., Derenko, M., Malyarchuk, B., Dambueva, I., Grzybowski, T. & Miścicka-Śliwka, D. Allelic and haplotypic frequencies at 11 Y-STR loci in Buryats from South-East Siberia. Forensic Sci. Int. 164, 271–275 (2006).
Rogalla, U., Woźniak, M., Swobodziński, J., Derenko, M., Malyarchuk, B. A., Dambueva, I. et al. A novel multiplex assay amplifying 13 Y-STRs characterized by rapid and moderate mutation rate. Forensic Sci. Int. Genet. 15, 49–55 (2015).
Kittler, R., Erler, A., Brauer, S., Stoneking, M. & Kayser, M. Apparent intrachromosomal exchange on the human Y chromosome explained by population history. Eur. J. Hum. Genet. 11, 304–314 (2003).
Park, M. J., Shin, K.-J., Kim, N. Y., Yang, W. I., Cho, S.-H. & Lee, H. Y. Characterization of deletions in the DYS385 flanking region and null alleles associated with AZFc microdeletions in Koreans. J. Forensic Sci. 53, 331–334 (2008).
Dulik, M. C., Osipova, L. P. & Schurr, T. G. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS ONE 6, e17548 (2011).
He, J. & Guo, F. Population genetics of 17 Y-STR loci in Chinese Manchu population from Liaoning Province, Northeast China. Forensic Sci. Int. Genet. 7, e84–e85 (2013).
Zhu, B., Wu, Y., Shen, C., Yang, T., Deng, Y., Xun, X. et al. Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic group residing in Qinghai province of China. Forensic Sci. Int. 175, 238–243 (2008).
Derenko, M., Malyarchuk, B., Denisova, G. A., Woźniak, M., Dambueva, I., Dorzhu, C. et al. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum. Genet. 118, 591–604 (2006).
Derenko, M., Malyarchuk, B., Denisova, G. A., Woźniak, M., Grzybowski, T., Dambueva, I. et al. Y-chromosome haplogroup N dispersals from south Siberia to Europe. J. Hum. Genet. 52, 763–770 (2007).
Malyarchuk, B., Derenko, M., Denisova, G., Maksimov, A., Woźniak, M., Grzybowski, T. et al. Ancient links between Siberians and Native Americans revealed by subtyping the Y chromosome haplogroup Q1a. J. Hum. Genet. 56, 583–588 (2011).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139, 457–462 (1995).
Willuweit, S. & Roewer, L. The new Y chromosome haplotype reference database. Forensic Sci. Int. Genet. 15, 43–48 (2015).
Zhivotovsky, L. A., Underhill, P. A., Cinnioglu, C., Kayser, M., Morar, B., Kivisild, T. et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74, 50–61 (2004).
Ballantyne, K. N., Goedbloed, M., Fang, R., Schaap, O., Lao, O., Wollstein, A. et al. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am. J. Hum. Genet. 87, 341–353 (2010).
Matsumura, S. & Forster, P. Generation time and effective population size in Polar Eskimos. Proc. Biol. Sci. 275, 1501–1508 (2008).
Goedbloed, M., Vermeulen, M., Fang, R. N., Lembring, M., Wollstein, A., Ballantyne, K. et al. Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR Yfiler PCR amplification kit. Int. J. Legal Med. 123, 471–482 (2009).
Hallast, P., Batini, C., Zadik, D., Maisano Delser, P., Wetton, J. H., Arroyo-Pardo, E. et al. The Y-chromosome tree bursts into leaf: 13,000 high-confidence SNPs covering the majority of known clades. Mol. Biol. Evol. 32, 661–673 (2015).
Bandelt, H.-J., Macaulay, V. & Richards, M. Median networks: speedy construction and greedy reduction, one simulation, and two case studies from human mtDNA. Mol. Phylogenet. Evol. 16, 8–28 (2000).
Wierdl, M., Dominska, M. & Petes, T. D. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics 146, 768–779 (1997).
Kayser, M., Kittler, R., Erler, A., Hedman, M., Lee, A. C., Mohyuddin, A. et al. A comprehensive survey of human Y-chromosomal microsatellites. Am. J. Hum. Genet. 74, 1183–1197 (2004).
Pugach, I., Matveev, R., Spitsyn, V., Makarov, S., Novgorodov, I., Osakovsky, V. et al. The complex admixture history and recent southern origins of Siberian populations. bioRxiv http://dx.doi.org/10.1101/018770 (2015).
Shubin, A. C. Brief Essay of Ethnic History of the Evenks from Trans-Baikal Region (XVIII-XX centries) (Buryat Publishing House, Ulan-Ude, Russia, 1973).
Buraev, I. D. & Shagdarov, L. D. in The Buryats (eds Abaeva, L. L. & Zhukovskaja, N. L.) 227–250 (Nauka, Moscow, Russia, 2004).
Dashibalov, B. B. Archaeological Sites of Kurykan and Khori (Buryat Scientific Center of SBRAS, Ulan-Ude, Russia, 1995).
Acknowledgements
This research was financially supported by the Program of Presidium of Russian Academy of Sciences ‘Basic research for the development of the Russian Arctic’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Malyarchuk, B., Derenko, M., Denisova, G. et al. Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407. J Hum Genet 61, 491–496 (2016). https://doi.org/10.1038/jhg.2016.14
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.14
This article is cited by
-
Mitogenomics of modern Mongolic-speaking populations
Molecular Genetics and Genomics (2022)
-
Phylogenetic analysis of the Y-chromosome haplogroup C2b-F1067, a dominant paternal lineage in Eastern Eurasia
Journal of Human Genetics (2020)
-
Mitogenomic diversity and differentiation of the Buryats
Journal of Human Genetics (2018)
-
Whole sequence analysis indicates a recent southern origin of Mongolian Y-chromosome C2c1a1a1-M407
Molecular Genetics and Genomics (2018)