Abstract
Takayasu arteritis (TAK) is an immune-mediated vasculitis affecting large arteries first reported in 1908 from Japan. Case reports of familial onset of TAK from Japan and other countries indicated genetic contribution to TAK onset beyond ethnicity. Genetic studies of TAK have been performed mainly addressing the human leukocyte antigen (HLA) locus. HLA genetic studies of TAK that have previously been reported are reviewed in this manuscript. HLA-B*52:01 is associated with TAK beyond population. Many of the associations other than HLA-B*52:01 can be explained by a haplotype with HLA-B*52:01. HLA-B*67:01 is a novel susceptibility HLA-B allele to TAK confirmed in the Japanese population. Further independent associations are suggested in the HLA locus. Involvement of the 171st and 67th amino acid residues with TAK onset has been indicated. The 67th amino acid may explain the difference in susceptibility effects to TAK and Behçet’s disease between HLA-B*52:01 and *51:01. HLA-B*52:01 is associated not only with TAK susceptibility but also with clinical phenotypes. Recent genome-wide association studies of TAK revealed multiple non-HLA susceptibility genes. In particular, the IL12B region seems to have a central role in TAK onset and its progression. Whether TAK and giant cell arteritis (GCA), the other vasculitis affecting large arteries, are the same disease is an interesting question to address in spite of different clinical manifestations between the two diseases. GCA is associated with HLA-DR4, which is not associated with TAK. GCA is not associated with HLA-Bw52. These two diseases seem not to share non-HLA susceptibility loci based on the recent genetic studies.
Similar content being viewed by others
Introduction
Takayasu arteritis (TAK) is one of the two vasculitis affecting the aorta and its large branches,1, 2 the other being giant cell arteritis (GCA).3 TAK was first reported from Japan in 1908 by Dr Mikito Takayasu, although there are other previous suspected case reports outside Japan.4 TAK is a relatively rare disease and the distribution is primarily in Asia. Japan is one of the countries with a high prevalence of TAK, estimated to be 0.01%.5 China, India, Turkey, Mexico and South America also show a relatively high prevalence of this disease. Although TAK is now classified as an autoimmune disease, it was only in the 1960s when researchers started regarding this disease as an immune-related disease. In 1962, Judge et al.6 suggested the involvement of autoimmunopathy with TAK based on four cases. In 1965, Riehl et al.7 reported that TAK is an autoimmune disease based on pathological findings. In 1967, Nakao et al.8 summarized 84 Japanese cases and analyzed the immunological aspects of 7 out of the 84 cases.
Because the prevalence of TAK is relatively low, it is not easy to find the familial history of TAK. It was in the 1960s when case reports of familial onset of TAK were started to be made.9 Although familial aggregation was reported mainly from Japan,10 there were also reports from outside Japan.11 These reports suggested genetic contributions in the susceptibility to TAK. On the basis of the evidence of heritability in TAK, researchers started the search to identify genetic factors associated with TAK. In this manuscript, genetic studies of TAK especially in the human leukocyte antigen (HLA) locus are reviewed.
HLA region
The summary of genetic studies for the HLA locus is shown in Table 1. Studies with more than 30 cases were chosen and sorted according to the publication year. Because more than half of the studies were reported from Japan, studies were divided into two groups: those from Japan and those from outside Japan.
HLA studies in Japan
The genetic study of TAK was launched in 1978 with a manuscript by Isohisa et al.,12 in which they focused on the HLA locus and genotyped the HLA-B region (Table 1). The HLA locus encodes the MHC protein which is composed of Class I and II molecules to present antigen to immune cells. MHC protein has a central role in self/non-self recognition, and the HLA locus has been known to have strong genetic associations with autoimmune diseases. Because TAK was recognized as an autoimmune disease, focusing on the HLA locus seemed to be a reasonable approach to identify susceptibility genes. Isohisa et al. found an increased ratio of HLA-B5 in the 65 patients of their study. Because HLA-B5 is composed mainly of two serotypes, namely HLA-Bw51 and Bw52, they further genotyped the two serotypes and found enrichment of Bw52 in the cases. Numano et al.,10 in 1979, genotyped six families with familial history of TAK and 65 patients and 128 controls for the HLA-A and B loci (Table 1). The samples from the 65 cases and 128 controls appear to be the same samples used by Isohisa et al.12 In this study, the authors reported that three parents out of 10 families with familial history of TAK were first cousins. They mentioned that the HLA-B5 made a haplotype with HLA-A9 and the haplotype was frequently observed in the familial cases. HLA-A10 was significantly increased in patients with TAK in the population study. They also mentioned suggestive decrease of Bw35 in the case samples. Sasazuki et al.13 expanded the results of Numano et al. and Isohisa et al. by analyzing the HLA-D locus. They found an association between TAK and HLA-DHO. In 1982, Isohisa et al.14 genotyped the HLA-A, B and D loci at the same time using 76 patients and 128 controls. They found that a haplotype of A9-Bw52-DHO was associated with TAK. In 1983, Numano et al.15 extended their previous studies by analyzing the HLA-DR, MT and MB loci. As a result, they did not find any significant associations in 52 patients with TAK. In 1982, Moriuchi et al.16 reported the association of HLA-Bw52, DR2 and MB1 with TAK.
Genotyping methods in most of the studies published before the 1990s adopted serological genotyping, which is different from those of the more recent studies. Thus, the correspondence between serotypes and sequence-based genotypes should be noted. HLA-A9 corresponds to HLA-A24 and A23 serotypes and mainly to HLA-A*24:02 genotype. HLA-A10 corresponds to HLA-A*26:01 genotype. HLA-Bw52 serotype basically corresponds to HLA-B*52:01 genotype. HLA-DR15 and DR16 compose HLA-DR2. Because the prevalence of TAK is low even in Japan, it was hard to accumulate a big number of DNA samples from patients with TAK. In the 1970s and 80s, it was not easy to collect a large number of control DNA samples as well. Thus, we should be cautious with the published data because two independent reports could be using the same samples. In addition, careful evaluation of all the genetic studies is necessary with regards to how many HLA regions were genotyped. From these points of view, we should note that the samples in the studies by Numano, Isohisa and Sasazuki, including controls as well as cases, seemed to largely overlap, although the authors did not always clearly mention this point in their manuscripts. We should therefore not regard their studies as purely replication studies of their previous reports, but expansion of loci and haplotype using the same or very similar samples. However, the study by Moriuchi et al.16 is an independent study in the Japanese population, and hence this work should be regarded as a replication study of HLA-B52. Because HLA-B*52:01 is in strong linkage disequilibrium (LD) with HLA-DRB1*15:02 in the Japanese population, the association of DR2 reported by Moriuchi et al. should be explained by HLA-DRB1*15:02 in LD with HLA-B*52:01, indicating one association. The lack of association in DRB1 in the report by Numano et al.15 in 1983 seemed contradictive to the reports by Isohisa et al. as well as their previous reports. However, HLA-DR2 showed a trend of enrichment in TAK patients with a relative risk of 1.52 in their study. Although the effect size was much smaller than that observed in the report by Moriuchi et al., the lack of the association in DR2 might be explained by the lack of power in the analysis, and the results in Numano et al. were not contradictory to previous reports. While HLA-B*52:01 is strongly linked with HLA-A*24:02 (A9 serum antigen), A9 is also tagged with other HLA-B alleles. This should be the reason why Numano et al. found an association for A9 only in the familial analysis and not in the population study. Whereas Numano et al.10 reported a positive association of A10 with TAK, Isohisa et al.14 did not report the association. The suggestive protective association of Bw35 could be due to an inverse association of B*52:01. HLA-MB corresponds to HLA-DQ. Because HLA-MB1 corresponds to HLA-DQw1 and HLA-DQw1 was later split into HLA-DQw5 and DQw6, the association between TAK and MB1 by Moriuchi et al. should be explained by the linkage between B*52:01 and DQB1*06:01. This was later confirmed by Takeuchi et al.17 in 1990, when they found an association of DQw6 using 32 cases and 34 controls for genotyping the HLA-D region. The association of HLA-DHO was shown to be dependent on the association of HLA-Bw52 in the report by Isohisa et al.14 Thus, the associations reported before 1990 could be explained by HLA-Bw52/HLA-B*52:01, namely, LD or a haplotype between HLA-Bw52/HLA-B*52:01 and A9/A24/A*24:02, DR2/DR15/DRB1*15:02 and/or MB1/DQw6/DQB1*06:01.
Dong et al.18 genotyped a total of 64 cases and 317 controls for the HLA-A, B, C, DR, DQ and DP regions in 1992. This study had a large number of controls and they genotyped as many as six loci simultaneously. They found HLA-Bw52, DRB1*15:02, DRB5*01:02, DQA1*01:03, DQB1*06:01 and DPB1*09:01 as the susceptibility alleles which made a haplotype. They also found HLA-Bw54, DRB1*04:05, DRB4*01:01, DQA1*03:01, DQB1*04:01 as the protective alleles which made another haplotype. Yoshida et al.19 in 1993 genotyped 64 cases and 156 controls for HLA-B and reported that HLA-B39.2 as well as HLA-B52 was associated with TAK. In 1996, Kimura et al.20 analyzed a total of 138 cases and 492 controls for the HLA-B, HLA-DRB1, DQA1 and DPB1 alleles, and found associations in HLA-B52 and B39. This study was the largest study in the world genotyping HLA-B, until 2012 when a Turkish team genotyped 330 cases21 and the largest study in Japan until 2013 when we reported the results of 173 patients.5 The samples used in these three studies by Dong, Yoshida and Kimura seemed to overlap. However, because this group was an independent group from the groups of Numano and Moriuchi, the association of HLA-B*52:01 in the Japanese population was again confirmed. Kimura et al. expanded their studies to other regions. In 1998, Kimura et al.22 genotyped the samples for the MICA region and found that MICA is associated with TAK. In 2000, Kimura et al.23 further genotyped microsatellite markers around HLA-B, HLA-C, MICA and MICB, namely, C1-2-A, MIB, C1-4-1, C1-2-5 and C1-3-1. They found positive associations in C1-2-A, MIB, C1-2-5 and C1-3-1. Shibata et al.,24 in the same group, genotyped the NFKBIL1 region and found an association in this locus in 2006. Recently, An et al.,25 in the same group, reported that inhibitor of kB-like protein (IkBL), encoded by NFKBIL1, regulates alternative splicing of immune-related genes.
Takamura et al.26 in 2012 showed that HLA-B*67:01 as well as HLA-B*52:01 are associated with TAK using 97 patients and 371 controls. This group is a novel group to analyze the genetic architecture of TAK. They genotyped HLA-A, C, DRB1 and DQB1 in addition to HLA-B, but did not find other associations. Our group in Kyoto University started to collect DNA samples of TAK and also independently showed HLA-B*67:01 as a susceptibility HLA-B allele to TAK using 173 patients of TAK and 2000 controls.5 Thus, at least five independent groups showed the association between HLA-B52 (namely, B*52:01) and TAK. Although previous studies showed that HLA-B39 was associated with TAK, recent studies could not confirm the association. Although there are several loci other than HLA-B reported to be associated with TAK, replication studies are essential to show convincing results. We also reported a genome-wide association study (GWAS) of TAK in 2013,27 where we found that a single nucleotide polymorphism (SNP) in strong LD with HLA-B*52:01 showed the strongest association among the HLA loci. When we conditioned the associations in the HLA locus for the SNP, we still found the suggestive associations, suggesting further independent associations in the HLA locus. Because the associations were not in LD with HLA-B*67:01 (unpublished data), there should be other associations aside from HLA-B*52:01 and *67:01.
HLA studies outside Japan
Genetic studies outside Japan using a relatively big number of cases started to be published in the 1990s. In 1982, Volkman et al.,28 an American group, reported the association between TAK and DR4 and MB3 using 11 patients of TAK. Khraishi et al.29 reported in 1992 a genetic study from North America in which they compared 21 cases and 243 controls for 73 class I antigens and 13 class II antigens and did not find any differences. They failed to find subjects carrying the B52 antigen, but they did not present the detailed results for class I antigens. It might be noted that they did not find even a tendency of enrichment of DR2 in their patients. In 1982, Castro et al.,30 a Mexican group, also reported the association of HLA-B5 with TAK, suggesting the association of HLA-B5 beyond ethnicity. Although other Mexican groups have published genetic studies for TAK, the number in each study has been quite limited.31 Girona et al.32 reported the results of genotyping 12 samples for HLA-A, B, C, DR and DQ. They found that HLA-B62 and DR6 were associated with TAK, and they suggested that the association of DR6 could be explained by DRB1*13:01. Soto et al.33 genotyped a total of 40 Mexican cases and reported associations of HLA-B39, B44 and B52 in 2006, and confirmed that B52 is associated with TAK. In 2000, Salazar et al.34 genotyped 16 Colombian patients with TAK for HLA class I and II. They did not find associations of B52 and DRB1*13:01, but found associations of DRB1*16:02 and *10:01. Satter et al.35 reported that four Arabic patients with TAK had Bw35, A2, A9 and DR7. It might be noted that three out of the four patients had DR2. An Indian genetic study was first reported in 1991 by Rose et al.36 using 50 cases. They found associations in HLA-B5 and B21 as susceptibility alleles and A19 as a protective allele.36 Later, Mehra et al.37 increased the number of cases to 80 samples and confirmed the association of B5, but they failed to confirm the associations of B21 and A19. They performed genotyping using a fraction of their cases and controls who were positive for B5, and showed that the frequency of B51 and B52 among these individuals did not seem to be different in the cases and in the controls, suggesting that both B51 and B52 were associated with TAK. Park and Park38 reported in 1992 the genetic association results for TAK in the Korean population. They showed that HLA-B52, Cw6, DR7 and DQw2 are associated with TAK. DR7 and DQw2 are not in LD with B52. Lee et al.39 reported another Korean study in 2007 where they found suggestive associations of A*30:01, B*52:01 and DRB1*15:02. The samples used in the two studies did not overlap, thus the association of B52 should be convincing in Korea. The association of DRB1*15:02 should be explained by B52. Although Lv et al.40 showed associations of HLA-DPB1*09 and 17:01 with TAK in the Han Chinese population, they did not provide data of HLA-B. Because HLA-DP*09 is strongly associated with HLA-DRB1*15:02, the association of DPB1*09 could be explained by DRB1*15:02 in LD with B*52:01. Because DPB1*17:01 is very rare in Japan, it is difficult to assume how the association should be attributed. The same group recently reported the associations of DRB1*07.41 It should be noted that the association of DRB1*07 in TAK was also reported by the group from Korea, and hence this may suggest another association in the DRB1 locus in East Asian populations. Sahin et al.21 genotyped 330 Turkish patients and 210 healthy controls for HLA-B51 and B52 by allele-specific PCR. They found the significant association between B52 and TAK, but not between B51 and TAK. Karageorgaki et al.42 analyzed a total of 44 cases in Greece and genotyped 22 cases for HLA-B in 2009, where they confirmed that B52 is associated with TAK.
Whereas the association of HLA-B52 is not apparent in North American populations, Saruhan-Direnskeneli et al.43 showed GWAS of TAK and they found a strong association signal in the HLA-B region in European Americans. HLA-B imputation showed a strong association of HLA-B*52:01. Although the most significant SNP did not strongly tag HLA-B*52:01, their imputation results showed that HLA-B*52:01 is associated with TAK in North Americans. When they adjusted for the top SNP, they still observed a significant association signal between the HLAB/HLA-DRB1 loci.
These results indicate that HLA-B*52:01 is associated with TAK beyond ethnicity, including Asians, Turkish and Americans and that other susceptibility alleles or genetic loci should exist in the HLA locus in addition to HLA-B*52:01. No alleles other than B*52:01 were established to be associated with TAK beyond population.
Associations between amino acid residues and susceptibility to TAK
Yoshida et al.19 in 1993 mentioned common amino acid residues, namely the 63rd glutamic acid and the 67th serine as important based on the common amino acid residues between HLA-B39.2 and the B52 protein. Vargas-Alarcon et al.44 analyzed 26 Mexican cases and 62 controls in 2005 and they showed that the 63rd glutamic acid and the 67th serine might be involved with susceptibility to TAK. However, enough power is required for these kinds of analyses to be conclusive. In 2013, we also showed that two amino acid residues, namely, positions 171 and 67, are important for TAK susceptibility using 173 cases and 2000 controls.5 Because the 67th amino acid is one of the two amino acid residues that differ between HLA-B*51:01 and 52:01 protein, our findings may explain why HLA-B*52:01 is associated with TAK and HLA-B*51:01 is not. As the 171st amino acid residue is characterized by the B*52:01 protein, the involvement of the 171st and 67th amino acid residues should be convincing across different populations. Because both amino acid residues reside at peptide binding grooves, the associations suggest that a common peptide recognized by these positions could be important for the development of this disease.
Associations between clinical manifestation and HLA alleles
The association between HLA alleles and clinical manifestations in TAK has also been addressed in the context of TAK-associated HLA alleles. Yajima et al.,45 in 1996, analyzed the association between HLA-Bw52 and aortic regurgitation (AR) in 87 patients. They reported higher complication of AR in patients with Bw52 than patients without. Kitamura et al.46 showed that the HLA-B*52:01 is associated with AR, ischemic heart disease and pulmonary infarction. HLA-B*39:01 was shown to be associated with renal stenosis.46 As patients’ data used by Kitamura et al. are independent from that of Yajima’s group, the association between HLA-B*52:01 and AR should be regarded as replicated. Takamura et al.26 analyzed the associations between B52 or B67 and age at disease onset, distribution of arteritis, pulmonary involvement, AR, systemic hypertension, steroid resistance and recurrence rate in TAK. Although they did not find significant associations, they found tendencies that AR, steroid resistance and recurrence in TAK were higher in patients carrying HLA-B52. Thus, the association between HLA-B*52:01 and AR should be convincing. The associations between HLA-B*52:01 and other phenotypes should be replicated. Our group analyzed the associations between HLA-B*67:01 and age at onset, female ratio, TAK classifications and AR,5 but we did not find significant associations. However, based on the frequency of HLA-B*67:01 in TAK (~5%), it is clear that both studies analyzing the associations of B*67:01 were underpowered.
Non-HLA region
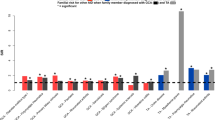
Although the association between the HLA locus and TAK susceptibility has been confirmed, genetic components outside HLA are still under investigation. GWAS has enabled us to elucidate the genetic background of complex diseases including autoimmune diseases.47, 48 Our group and another group in Turkey performed GWAS of TAK for the first time in the world. Both groups identified IL12B as a susceptibility locus to TAK. Our group showed another association in chr 17.27 The Turkish group showed FCGR2A/3A as another susceptibility locus outside HLA,49 and they recently published a follow-up study to show IL6, RPS9/LILRB3 and chr21q22 as susceptibility regions to TAK.50 However, the associations in IL6 and RPS9/LILRB3 showed borderline significance, and hence intensive replication studies are favorable. As tocilizumab, a monoclonal antibody against IL6 receptor, is reported to be effective to TAK;51 the IL6 region as a susceptibility region to TAK is interesting.
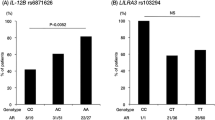
It should be notable that the susceptibility alleles to TAK seem to have high effect sizes (rs6871626 in IL12B in the Japanese study: odds ratio=1.75 (95% confidence interval:1.42–2.16)). Because genetic determinants to other autoimmune diseases identified from GWAS have relatively mild effect sizes (odds ratio:1.07~1.47 in rheumatoid arthritis),52, 53 the high effect sizes in TAK susceptibility genes may indicate that TAK patients studied seem to be more homogeneous than other complex diseases. This may suggest that whereas low prevalence of TAK makes it difficult to accumulate the number of DNA samples, diagnosis of TAK which is basically made by specialists, is accurate and the subjects were not contaminated by other diseases.
IL12B as a susceptibility region to TAK is implicative. IL12B encodes IL12p40, a subunit of IL12 and IL23. IL12B SNP showed a strong effect on TAK susceptibility in the Japanese.27 Furthermore, IL12B showed a synergistic effect on TAK susceptibility in combination with HLA-B*52:01.27 In addition, IL12B is associated with a higher complication rate of AR, severity of AR and high disease activity.27 Furthermore, a Japanese group recently showed that the IL12B SNP is associated with the combination of young onset, frequent relapse and poor response to treatment.54 IL12 is an essential cytokine for proliferation of Th1 cells55 and IL23 is deeply related to survival and activation of Th1756 and both of Th1 and Th17 are associated with autoimmune diseases. Higher IL12p40 levels are reported in patients with TAK compared with healthy population57 and our in-house data suggest that the risk allele of IL12B is associated with higher expression of IL12B and IL12p40 protein in patients with TAK (unpublished data). The importance of IL12B suggests dysregulation of Th1 and/or Th17 in patients with TAK owing to overproduction of IL12p40.
These results suggest that IL12B has a fundamental role on TAK development and progression. Thus, IL12B and IL12/23p40 are promising treatment targets in patients with TAK. Ustekinumab, a monoclonal antibody against IL12/23p40, is widely used for patients with Crohn’s disease, one of the inflammatory bowel diseases, and psoriasis and displayed favorable outcomes.58, 59 Our group has recently shown that ulcerative colitis, the other inflammatory bowel disease, is frequently found in patients with TAK,60 suggesting common molecular pathways underlying TAK and inflammatory bowel diseases. Inflammatory bowel diseases and psoriasis share IL12B as a susceptibility gene,61, 62, 63 and hence ustekinumab could also be effective for patients with TAK. In fact, when we performed a pilot clinical trial using ustekinumab to patients with refractory TAK for whom conventional treatment was not effective, all of the three patients showed good response to ustekinumab (Terao et al., Scand J Rheumatol in press). This indicates that ustekinumab could be a treatment option to refractory TAK. Further clarification of geneteic architectures of TAK would lead to the identification of further novel treatment options.
Comparison with GCA
Although clinical manifestations are different between TAK and GCA, there is controversy over whether or not they are the same disease entity. Comparison of genetic architecture between the two diseases may offer a clear answer.
Previous genetic studies by European groups addressing HLA alleles associated with GCA have shown an association of HLA-DR4, especially DRB1*04:01 and *04:04.64, 65, 66, 67, 68, 69 They did not find consistent associations in HLA Class I including the HLA-B locus.70, 71 Considering the prevalence of HLA-DR4, it is believed that Asian genetic studies will identify the same association, and a Japanese group is currently recruiting DNA subjects. This point would be addressed in the near future.
A Spanish group has recruited a large number of GCA samples in the European population. They performed candidate studies and reported that IL17A,72 PTPN2273 and NLRP174 are associated with GCA. However, the associations of these results were far from GWAS significance. The Spanish group has recently reported the results of GWAS.75 They found a significant association only in the HLA locus. Thus, there are no established non-HLA susceptibility loci to GCA. The top signal in the HLA locus tagged the histidine residue at amino acid position of 13 in HLA-DRB1 protein. An imputation of HLA classical alleles showed the strong association of HLA-DR4 with GCA. An independent protective association signal was observed in HLA-DQA1 after conditioning for the top signal in HLA-DRB1. After conditioning for the associations of HLA Class II locus, they still observed a significant association in HLA-B in a protective manner. Notably, the HLA-B alleles carrying the protective amino acid residue contain HLA-B*52:01. Thus, there are no evidence to date, supporting genetic similarities for non-HLA loci and the HLA locus between TAK and GCA. Comparison of results of two GWAS would lead to clarification of similarity and difference of genetic architectures including polygenetic architecture76 between the two diseases.
References
Terao, C., Yoshifuji, H. & Mimori, T. Recent advances in Takayasu arteritis. Int. J. Rheum. Dis. 17, 238–247 (2014).
Isobe, M. Takayasu arteritis revisited: Current diagnosis and treatment. Int. J. Cardiol. 168, 3–10 (2013).
Jennette, J. C., Falk, R. J., Bacon, P. A., Basu, N., Cid, M. C., Ferrario, F. et al. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum. 65, 1–11 (2013).
Terao, C. History of Takayasu arteritis and Dr. Mikito Takayasu. Int. J. Rheum. Dis 17, 931–935 (2014).
Terao, C., Yoshifuji, H., Ohmura, K., Murakami, K., Kawabata, D., Yurugi, K. et al. Association of Takayasu arteritis with HLA-B 67:01 and two amino acids in HLA-B protein. Rheumatology (Oxford) 52, 1769–1774 (2013).
Judge, R. D., Currier, R. D., Gracie, W. A. & Figley, M. M. Takayasu's arteritis and the aortic arch syndrome. Am. J. Med. 32, 379–392 (1962).
Riehl, J. L. & Brown, W. J. Takayasu's Arteritis: an auto immune disease. Arch. Neurol. 12, 92–97 (1965).
Nakao, K., Ikeda, M., Kimata, S., Niitani, H. & Niyahara, M. Takayasu's arteritis. Clinical report of eighty-four cases and immunological studies of seven cases. Circulation 35, 1141–1155 (1967).
Hirsch, M. S., Aikat, B. K. & Basu, A. K. Takayasu's Arteritis. Report of five cases with immunologic studies. Bull. Johns Hopkins Hosp. 115, 29–64 (1964).
Numano, F., Isohisa, I., Maezawa, H. & Juji, T. HL-A antigens in Takayasu's disease. Am. Heart J. 98, 153–159 (1979).
Hermann, V. B. & Pluhor, J. Beitrage zur pathogenese des Aortenbogen Syndrome. Zschr. Inn. Med. 10, 453 (1964).
Isohisa, I., Numano, F., Maezawa, H. & Sasazuki, T. HLA-Bw52 in Takayasu disease. Tissue Antigens 12, 246–248 (1978).
Sasazuki, T., Ohta, N., Isohisa, I., Numano, F. & Maezawa, H. Association between Takayasu disease and HLA-DHO. Tissue Antigens 14, 177–178 (1979).
Isohisa, I., Numano, F., Maezawa, H. & Sasazuki, T. Hereditary factors in Takayasu's disease. Angiology 33, 98–104 (1982).
Numano, F., Isohisa, I., Egami, M., Ohta, N. & Sasazuki, T. HLA-DR MT and MB antigens in Takayasu disease. Tissue Antigens 21, 208–212 (1983).
Moriuchi, J., Wakisaka, A., Aizawa, M., Yasuda, K., Yokota, A., Tanabe, T. et al. HLA-linked susceptibility gene of Takayasu Disease. Hum. Immunol. 4, 87–91 (1982).
Takeuchi, Y., Matsuki, K., Saito, Y., Sugimoto, T. & Juji, T. HLA-D region genomic polymorphism associated with Takayasu's arteritis. Angiology 41, 421–426 (1990).
Dong, R. P., Kimura, A., Numano, F., Yajima, M., Hashimoto, Y., Kishi, Y. et al. HLA-DP antigen and Takayasu arteritis. Tissue Antigens 39, 106–110 (1992).
Yoshida, M., Kimura, A., Katsuragi, K., Numano, F. & Sasazuki, T. DNA typing of HLA-B gene in Takayasu's arteritis. Tissue Antigens 42, 87–90 (1993).
Kimura, A., Kitamura, H., Date, Y. & Numano, F. Comprehensive analysis of HLA genes in Takayasu arteritis in Japan. Int. J. Cardiol. 54 ( (Suppl), ), S61–S69 (1996).
Sahin, Z., Bicakcigil, M., Aksu, K., Kamali, S., Akar, S., Onen, F. et al. Takayasu's arteritis is associated with HLA-B*52, but not with HLA-B*51, in Turkey. Arthritis Res. Ther. 14, R27 (2012).
Kimura, A., Kobayashi, Y., Takahashi, M., Ohbuchi, N., Kitamura, H., Nakamura, T. et al. MICA gene polymorphism in Takayasu's arteritis and Buerger's disease. Int. J. Cardiol. 66 (Suppl 1),, S107–S113 (1998).
Kimura, A., Ota, M., Katsuyama, Y., Ohbuchi, N., Takahashi, M., Kobayashi, Y. et al. Mapping of the HLA-linked genes controlling the susceptibility to Takayasu's arteritis. Int. J. Cardiol. 75 (Suppl 1),, S105–S110 (2000).
Shibata, H., Yasunami, M., Obuchi, N., Takahashi, M., Kobayashi, Y., Numano, F. et al. Direct determination of single nucleotide polymorphism haplotype of NFKBIL1 promoter polymorphism by DNA conformation analysis and its application to association study of chronic inflammatory diseases. Hum. Immunol. 67, 363–373 (2006).
An, J., Nakajima, T., Shibata, H., Arimura, T., Yasunami, M. & Kimura, A. A novel link of HLA locus to the regulation of immunity and infection: NFKBIL1 regulates alternative splicing of human immune-related genes and influenza virus M gene. J. Autoimmun. 47, 25–33 (2013).
Takamura, C., Ohhigashi, H., Ebana, Y. & Isobe, M. New human leukocyte antigen risk allele in Japanese patients with Takayasu arteritis. Circ. J. 76, 1697–1702 (2012).
Terao, C., Yoshifuji, H., Kimura, A., Matsumura, T., Ohmura, K., Takahashi, M. et al. Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the IL12B and HLA-B regions in a Japanese population. Am. J. Hum. Genet. 93, 289–297 (2013).
Volkman, D. J., Mann, D. L. & Fauci, A. S. Association between Takayasu's arteritis and a B-cell alloantigen in North Americans. N. Engl. J. Med. 306, 464–465 (1982).
Khraishi, M. M., Gladman, D. D., Dagenais, P., Fam, A. G. & Keystone, E. C. HLA antigens in North American patients with Takayasu arteritis. Arthritis Rheum. 35, 573–575 (1992).
Castro, G., Chavez-Peon, C., Sanchez-Torres, G. & Reyes, P. A. HLA A and B antigens in Takayasu's arteritis. Rev. Invest. Clin. 34, 15–17 (1982).
Vargas-Alarcon, G., Flores-Dominguez, C., Hernandez-Pacheco, G., Zuniga, J., Gamboa, R., Soto, M. E. et al. Immunogenetics and clinical aspects of Takayasu's arteritis patients in a Mexican Mestizo population. Clin. Exp. Rheumatol. 19, 439–443 (2001).
Girona, E., Yamamoto-Furusho, J. K., Cutino, T., Reyes, P., Vargas-Alarcon, G., Granados, J. et al. HLA-DR6 (possibly DRB1*1301) is associated with susceptibility to Takayasu arteritis in Mexicans. Heart Vessels 11, 277–280 (1996).
Soto, M. E., Vargas-Alarcon, G., Cicero-Sabido, R., Ramirez, E., Alvarez-Leon, E. & Reyes, P. A. Comparison distribution of HLA-B alleles in mexican patients with takayasu arteritis and tuberculosis. Hum. Immunol. 68, 449–453 (2007).
Salazar, M., Varela, A., Ramirez, L. A., Uribe, O., Vasquez, G., Egea, E. et al. Association of HLA-DRB1*1602 and DRB1*1001 with Takayasu arteritis in Colombian mestizos as markers of Amerindian ancestry. Int. J. Cardiol. 75 (Suppl 1),, S113–S116 (2000).
Sattar, M. A., White, A. G., Eklof, B. & Fenech, F. F. Takayasu's disease in Arabs. Postgrad. Med. J. 61, 387–390 (1985).
Rose, S., Mehra, N. K., Kumar, R. & Vaidya, M. C. HLA-B5 and B21 antigens in aortoarteritis. Indian J. Pediatr. 58, 85–89 (1991).
Mehra, N. K., Jaini, R., Balamurugan, A., Kanga, U., Prabhakaran, D., Jain, S. et al. Immunogenetic analysis of Takayasu arteritis in Indian patients. Int. J. Cardiol. 66 (Suppl 1),, S127–S132 (1998).
Park, M. H. & Park, Y. B. HLA typing of Takayasu arteritis in Korea. Heart Vessels Suppl. 7, 81–84 (1992).
Lee, S. W., Kwon, O. J., Park, M. C., Oh, H. B., Park, Y. B. & Lee, S. K. HLA alleles in Korean patients with Takayasu arteritis. Clin. Exp. Rheumatol. 25, S18–S22 (2007).
Lv, N., Dang, A., Wang, Z., Zheng, D. & Liu, G. Association of susceptibility to Takayasu arteritis in Chinese Han patients with HLA-DPB1. Hum. Immunol. 72, 893–896 (2011).
Lv, N., Wang, Z., Dang, A., Zhu, X., Liu, Y., Zheng, D. et al. HLA-DQA1, DQB1 and DRB1 alleles associated with Takayasu arteritis in the Chinese Han population. Hum. Immunol. 76, 241–244 (2015).
Karageorgaki, Z. T., Bertsias, G. K., Mavragani, C. P., Kritikos, H. D., Spyropoulou-Vlachou, M., Drosos, A. A. et al. Takayasu arteritis: epidemiological, clinical, and immunogenetic features in Greece. Clin. Exp. Rheumatol. 27, S33–S39 (2009).
Saruhan-Direskeneli, G., Hughes, T., Aksu, K., Keser, G., Coit, P., Aydin, S. Z. et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am. J. Hum. Genet. 93, 298–305 (2013).
Vargas-Alarcon, G., Hernandez-Pacheco, G., Soto, M. E., Murguia, L. E., Perez-Hernandez, N., Granados, J. et al. Comparative study of the residues 63 and 67 on the HLA-B molecule in patients with Takayasu's Arteritis. Immunol. Lett. 96, 225–229 (2005).
Yajima, M., Numano, F., Park, Y. B. & Sagar, S. Comparative studies of patients with Takayasu arteritis in Japan, Korea and India—ison of clinical manifestations, angiography and HLA-B antigen. Jpn. Circ. J. 58, 9–14 (1994).
Kitamura, H., Kobayashi, Y., Kimura, A. & Numano, F. Association of clinical manifestations with HLA-B alleles in Takayasu arteritis. Int. J. Cardiol. 66 (Suppl 1),, S121–S126 (1998).
Terao, C., Ohmura, K., Katayama, M., Takahashi, M., Kokubo, M., Diop, G. et al. Myelin basic protein as a novel genetic risk factor in rheumatoid arthritis—me-wide study combined with immunological analyses. PLoS ONE 6, e20457 (2011).
Terao, C., Yamada, R., Ohmura, K., Takahashi, M., Kawaguchi, T., Kochi, Y. et al. The human AIRE gene at chromosome 21q22 is a genetic determinant for the predisposition to rheumatoid arthritis in Japanese population. Hum. Mol. Genet. 20, 2680–2685 (2011).
Saruhan-Direskeneli, G., Hughes, T., Aksu, K., Keser, G., Coit, P., Aydin, S. Z. et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am. J. Hum. Genet. 67, 1361–1368 (2013).
Renauer, P., Saruhan-Direskeneli, G., Coit, P., Adler, A., Aksu, K., Keser, G. et al. Genome-wide association study identifies susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in Takayasu's arteritis. Arthritis Rheumatol. 67, 1361–1368 (2015).
Nakaoka, Y., Higuchi, K., Arita, Y., Otsuki, M., Yamamoto, K., Hashimoto-Kataoka, T. et al. Tocilizumab for the treatment of patients with refractory takayasu arteritis. Int. Heart J. 54, 405–411 (2013).
Kochi, Y., Okada, Y., Suzuki, A., Ikari, K., Terao, C., Takahashi, A. et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet. 42, 515–519 (2010).
Okada, Y., Terao, C., Ikari, K., Kochi, Y., Ohmura, K., Suzuki, A. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 44, 511–516 (2012).
Matsumura, T., Amiya, E., Tamura, N., Maejima, Y., Komuro, I. & Isobe, M. A novel susceptibility locus for Takayasu arteritis in the IL12B region can be a genetic marker of disease severity. Heart Vessels (e-pub ahead of print 18 March 2015).
Murphy, K. M. & Reiner, S. L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002).
Bettelli, E., Carrier, Y., Gao, W., Korn, T., Strom, T. B., Oukka, M. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Verma, D. K., Tripathy, N. K., Verma, N. S. & Tiwari, S. Interleukin 12 in Takayasu's arteritis: plasma concentrations and relationship with disease activity. J. Rheumatol. 32, 2361–2363 (2005).
Sandborn, W. J., Gasink, C., Gao, L. L., Blank, M. A., Johanns, J., Guzzo, C. et al. Ustekinumab induction and maintenance therapy in refractory Crohn's disease. N. Engl. J. Med. 367, 1519–1528 (2012).
Yeilding, N., Szapary, P., Brodmerkel, C., Benson, J., Plotnick, M., Zhou, H. et al. Development of the IL-12/23 antagonist ustekinumab in psoriasis: past, present, and future perspectives—ate. Ann. N. Y. Acad. Sci. 1263, 1–12 (2012).
Terao, C., Matsumura, T., Yoshifuji, H., Kirino, Y., Maejima, Y., Nakaoka, Y. et al. Takayasu arteritis and ulcerative colitis -high concurrence ratio and genetic overlap. Arthritis Rheumatol. (e-pub ahead of print 30 April 2015).
Jostins, L., Ripke, S., Weersma, R. K., Duerr, R. H., McGovern, D. P., Hui, K. Y. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Tsunemi, Y., Saeki, H., Nakamura, K., Sekiya, T., Hirai, K., Fujita, H. et al. Interleukin-12 p40 gene (IL12B) 3'-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 30, 161–166 (2002).
Cargill, M., Schrodi, S. J., Chang, M., Garcia, V. E., Brandon, R., Callis, K. P. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
Weyand, C. M., Hicok, K. C., Hunder, G. G. & Goronzy, J. J. The HLA-DRB1 locus as a genetic component in giant cell arteritis. Mapping of a disease-linked sequence motif to the antigen binding site of the HLA-DR molecule. J. Clin. Invest. 90, 2355–2361 (1992).
Weyand, C. M., Hunder, N. N., Hicok, K. C., Hunder, G. G. & Goronzy, J. J. HLA-DRB1 alleles in polymyalgia rheumatica, giant cell arteritis, and rheumatoid arthritis. Arthritis Rheum. 37, 514–520 (1994).
Rauzy, O., Fort, M., Nourhashemi, F., Alric, L., Juchet, H., Ecoiffier, M. et al. Relation between HLA DRB1 alleles and corticosteroid resistance in giant cell arteritis. Ann. Rheum. Dis. 57, 380–382 (1998).
Combe, B., Sany, J., Le Quellec, A., Clot, J. & Eliaou, J. F. Distribution of HLA-DRB1 alleles of patients with polymyalgia rheumatica and giant cell arteritis in a Mediterranean population. J. Rheumatol. 25, 94–98 (1998).
Dababneh, A., Gonzalez-Gay, M. A., Garcia-Porrua, C., Hajeer, A., Thomson, W. & Ollier, W. Giant cell arteritis and polymyalgia rheumatica can be differentiated by distinct patterns of HLA class II association. J. Rheumatol. 25, 2140–2145 (1998).
Jacobsen, S., Baslund, B., Madsen, H. O., Tvede, N., Svejgaard, A. & Garred, P. Mannose-binding lectin variant alleles and HLA-DR4 alleles are associated with giant cell arteritis. J. Rheumatol. 29, 2148–2153 (2002).
Armstrong, R. D., Behn, A., Myles, A., Panayi, G. S. & Welsh, K. I. Histocompatibility antigens in polymyalgia rheumatica and giant cell arteritis. J. Rheumatol. 10, 659–661 (1983).
Richardson, J. E., Gladman, D. D., Fam, A. & Keystone, E. C. HLA-DR4 in giant cell arteritis: association with polymyalgia rheumatica syndrome. Arthritis Rheum. 30, 1293–1297 (1987).
Marquez, A., Hernandez-Rodriguez, J., Cid, M. C., Solans, R., Castaneda, S., Fernandez-Contreras, M. E. et al. Influence of the IL17A locus in giant cell arteritis susceptibility. Ann. Rheum. Dis. 73, 1742–1745 (2014).
Serrano, A., Marquez, A., Mackie, S. L., Carmona, F. D., Solans, R., Miranda-Filloy, J. A. et al. Identification of the PTPN22 functional variant R620W as susceptibility genetic factor for giant cell arteritis. Ann. Rheum. Dis. 72, 1882–1886 (2013).
Serrano, A., Carmona, F. D., Castaneda, S., Solans, R., Hernandez-Rodriguez, J., Cid, M. C. et al. Evidence of association of the NLRP1 gene with giant cell arteritis. Ann. Rheum. Dis. 72, 628–630 (2013).
Carmona, F. D., Mackie, S. L., Martin, J. E., Taylor, J. C., Vaglio, A., Eyre, S. et al. A large-scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am. J. Hum. Genet. 96, 565–580 (2015).
International Schizophrenia Consortium., Purcell, S. M., Wray, N.R., Stone, J.L., Visscher, P.M., O'Donovan, M. C. et alInternational Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Terao, C. Revisited HLA and non-HLA genetics of Takayasu arteritis—where are we?. J Hum Genet 61, 27–32 (2016). https://doi.org/10.1038/jhg.2015.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.87
This article is cited by
-
Distinct gene signatures of monocytes and B cells in patients with giant cell arteritis: a longitudinal transcriptome analysis
Arthritis Research & Therapy (2023)
-
HLA-B52 allele in giant cell arteritis may indicate diffuse large-vessel vasculitis formation: a retrospective study
Arthritis Research & Therapy (2021)
-
Therapie der Takayasu-Arteriitis
Zeitschrift für Rheumatologie (2020)
-
Treatment of Giant Cell Arteritis and Takayasu Arteritis—Current and Future
Current Rheumatology Reports (2020)
-
Significant association between clinical characteristics and changes in peripheral immuno-phenotype in large vessel vasculitis
Arthritis Research & Therapy (2019)