Abstract
The term ‘genetics’ was coined before an understanding of DNA sequence data was achieved, and it is now insufficient to describe the broad areas in which DNA data have important roles. The term genomics is more broadly descriptive, but it does not provide a satisfactory conceptual framework that scientists can share. Here I propose a six-layer structure that describes the entire scientific field for ‘genomics’. The proposed layers are ‘life’ as the uppermost layer, followed by ‘species’, ‘population’, ‘family’, ‘individual’ and finally ‘cell’ as the bottommost layer. In each pair of adjacent layers, each member of the upper layer comprises a set of members of the lower layer. In each layer, we can define consistent partial orders of members based on genomic data in the forms of phylogenic and pedigree trees. Although total orders such as those defined for time and space in physics cannot be defined in biology, defining consistent partial orders allows mathematical analysis to be performed. I will show that mathematical genetics studies can be understood as attempts to bridge gaps between layers of the proposed six-layer structure, while genetic tests can be understood as procedures to differentiate among members of each layer by using genomic data.
Similar content being viewed by others
Six-layer structure for genomics
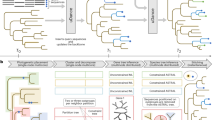
The scientific term ‘genetics’ originated just after the rediscovery of Mendel’s laws of inheritance.1 Thus, Bateson used, for the first time, the word ‘genetics’ in his letter to Cambridge University authorities in 1905, and used this word again at the third International Plant Breeding Congress in London in 1906.1 According to his definition, ‘genetics’ includes ‘heredity’ and ‘variation’.1 The use of the word ‘genetics’ subsequently increased along with a decrease in the use of the word ‘heredity’ (Figure 1). Recently, the use of DNA sequence data has become widespread and the term ‘genetics’, which was defined before the existence of DNA was known, has become inadequate to describe broad areas of science and medicine where DNA sequence data are used. For example, genomic changes in somatic and cancer cells are not necessarily covered by the concept ‘genetics’. If ‘genetics’ is used for the fields where genes are involved (gene-tics), variations in the genomes or DNA outside the genes are not covered by this concept. Recently, however, the use of the word ‘genomics’ has increased along with a decrease in the use of the word ‘genetics’ (Figure 1). The word ‘genomics’ was initially used to describe the field of biology dealing with DNA sequence data for the complete set of DNA within a single cell of an organism (genome). However, this term has been conceptually extended to include medical fields such as pharmacogenomics, cancer genomics and public health genomics. As the term ‘genetics’ no longer sufficiently describes all the main biological and medical fields in which DNA sequence data have important roles, a new term is needed and the term ‘genomics’ may fit this purpose. Currently, biological scientists and medical professionals are limited by the absence of a structured conceptual framework for sharing information. I propose here a conceptual framework named the ‘six-layer structure for genomics’ (Figure 2) to provide an extended concept of genomics that spans wide areas of biology and medicine.
Yearly changes of the frequencies of the words ‘heredity’, ‘genetics’ and ‘genomics’ in Google Books as shown by Ngram viewer (https://books.google.com/ngrams). The setting used for searches was ‘case insensitive’.
A series of six layers forms this structure. In each pair of adjacent layers, a member of the upper layer comprises a set of the members of the lower layer (Figure 2). Thus, the uppermost layer, ‘life’, is a set of ‘species’ (second layer), a species is a set of ‘populations’ (third layer) and so on forth; a ‘population is a set of ‘families’, a ‘family’ is a set of ‘individuals’ and an ‘individual’ is a set of ‘cells’. In my opinion, Darwin’s theory of evolution theoretically bridged the gap between the ‘life’ and ‘species’ layers, while Mendel’s laws theoretically connected the ‘family’ and ‘individual’ layers (Figure 2). When Darwin’s theory of evolution and Mendel’s laws of inheritance became known to the scientific community at the beginning of the twentieth century, the gap between these two theories was not easily bridged theoretically. This gap was later bridged by other works including those of population geneticists such as Wright, Haldane and Fisher, which led to the integrated concept of ‘modern evolutionary synthesis’ or ‘Neo-Darwinism’ of the 1930s and 1940s.2 I propose that these works effectively inserted the ‘population’ layer between the ‘species’ and the ‘family’ layers (Figure 2), and the modern integrated concept is widely accepted by the current scientific community. I do not propose a ‘molecule’ layer below the ‘cell’ layer, because molecules do not have genomes. I admit that the definitions of population and family layers are not as distinct as the other layers as the borders of families and populations are not as distinct as those of species, individuals and cells. However, the insertion of population and family layers benefits us to understand the roles of genomic data in various scientific and medical situations.
Application of mathematics in each of the layers
The proposed six-layer structure is useful for understanding the valid application of mathematics to genomics. It is clear that mathematics can be used powerfully in physics, but this has been less clear for biology. My opinion is that this difference exists because many objects of study in physics are totally ordered, but those in biology are not. Totally ordered set is defined as a set in which the orders of all pairs of the members are defined (equality may be allowed). For example, time and space, which are important objects of study in physics, comprise totally ordered sets. Mathematics using real numbers therefore fits the objects of physics very well. The importance of having well-ordered objects for the application of mathematics is shown by two simple examples (Figure 3). If the mass of a single object is found to be larger at time 1 than at time 0, we can say that the mass of the object increased with time (Figure 3a), whereas the reverse is true if the mass of the object is larger at time 0 than at time 1 (Figure 3b). The next example concerns the abundances of two different proteins in a biological system (Figures 3c and d). Imagine that we observed that the abundances of proteins X and Y decreased simultaneously. If the order of the effect is from X to Y (Figure 3c), we can expect that a drug that suppresses X will result in a decrease in Y. On the contrary, if the effect is from Y to X, the suppression of X often results in an increase in Y because of the feedback inhibition of Y when X increases (Figure 3d). Those two examples show that a consistent ordering of the objects of study is essential for the application of mathematics to real-world research. As many objects of study are totally ordered in physics, differential and integral can be applied (of course additional assumptions are necessary). In biology, the objects of study are not totally ordered, but we can find consistent orders in each of the six layers. Thus, in the ‘species’ layer (a set of species) we can construct a phylogenic tree based on the genomic sequences of various species (Figure 2), and in the ‘cell’ layer a reliable phylogenic tree can be constructed, as all cells in an animal body are derived from a single fertilized egg (Figure 2). For both of these layers, consistent partial orders can be defined, as most of their members have ancestors as defined by the phylogenic trees. Partially ordered set is defined as a set in which the orders of some (but not all) pairs of the members are defined (equality may be allowed). In the ‘individual’ layer, a pedigree tree defines the ordering, as each individual has a mother and a father (in species with sexual reproduction; Figure 2). Thus, we can define partial orders for each of the layers.
Simple examples showing the importance of having consistent orders of objects. (a, b) Change of the mass of an object from time 0 to time 1. If the order of these times is reversed, the conclusion is also reversed. (c, d) Changes in the abundances of proteins X and Y. If simultaneous changes in the abundances of proteins X and Y are observed, the effect of the suppression of X on Y differs depending on the direction of the effect between X and Y. If the direction of the effect is from Y to X, an increase in the abundance of Y often results in a decrease in the abundance of X because of inhibitory feedback mechanisms.
Genetic studies using mathematics
In the context of the proposed six-layer structure, many genetic studies can be understood as methods to bridge gaps between different layers based on the partial orders of the objects in those layers. For example, studies on molecular evolution can be viewed as attempts to bridge the gap between the ‘life’ and the ‘species’ layers by applying mathematics to the phylogeny of various species based on the orders defined by phylogenic trees (Figure 4a). Analyses of population structure by using single-nucleotide polymorphism data3 can be considered to be attempts to bridge the gap between the ‘species’ and the ‘population’ layers (Figure 4a). Linkage analysis is a procedure to bridge the gap between the ‘family’ and the ‘individual’ layers by using the orders defined by pedigree trees (Figure 4a). Genome-wide association studies (GWASs) are attempts to bridge the gap between the ‘population’ and the ‘individual’ layers (Figure 4a). In GWASs, the structuring of the population is a major problem, as the ‘family’ layer that exists between the ‘population’ and the ‘individual’ layers is skipped (Figure 4a). Studies on the application of mathematics to bridge the gap between the ‘individual’ and the ‘cell’ layers (Figure 4a) have begun.4 As all the cells in an animal body are derived from a single fertilized egg, we can define a partially ordered set in the ‘cell’ layer based on the somatic cell phylogeny (Figure 4a). This structure is quite similar to that of the ‘species’ layer, and equivalent mathematical procedures to those used in studies on molecular evolution are expected to also be applicable to the ‘cell’ layer.
An integrated understanding of various genetic studies and tests. (a) Genetic studies using mathematics can be understood as attempts to bridge the gap between two layers of the six-layer structure for genomics. (b) Genetic tests are often understood to be procedures to differentiate among members of one of the six layers by genomic sequences. DTC, direct to consumer; NIPT, non-invasive prenatal testing.
Various genetic tests can be understood in an integrated way
On the basis of the six-layer structure, many genetic tests can be understood as procedures to differentiate the members of a layer based on their genomic sequences (Figure 4b). A genetic test is present even in the ‘life’ layer. Astronomers are eager to discover life on an extraterrestrial planet, such as Mars (Figure 4b). This search can be interpreted as a genetic test to differentiate between life and non-life based on the presence or absence of a genomic sequence (Figure 4b). Clinical genetic tests for infection or metagenomic tests can be interpreted as genetic tests that differentiate among different species based on their genomic sequences (Figure 4b). Separation of clusters by principal component analysis using numerous single-nucleotide polymorphism genotypes can be defined as a genetic test for ethnicities (Figure 4b), and the diagnosis of genetic diseases can be interpreted as genetic tests in the ‘family’ and the ‘individual’ layers (Figure 4b).
Finding analogies between different layers may lead to major discoveries
We may consider phenotypes in each layer. For example, cancer, size and reaction to a compound are phenotypes in cell layer, while shape, size and presence of a certain protein are phenotypes of population and species layers. Association between genomic sequences and phenotypes in each layer are the targets of important studies in genomics. Considering the phenotypes in each layer, it may be possible to make major discoveries by finding analogies between different layers and extrapolating knowledge from one layer to another (trans-layer extrapolation). An example is our discovery of methylthioadenosine phosphorylase (MTAP) deficiency in human leukemias in 1982, which was the first discovery of a biochemical result of a mutation in a tumor suppressor gene in humans.5 This discovery was made possible by extrapolating knowledge about various genetic enzyme deficiencies in the ‘family’ and the ‘species’ layers to the ‘cell’ layer. As many genetic enzyme deficiencies can occur in humans (acting at the ‘family’ and the ‘individual’ layers) and as all humans are deficient in urate oxidase (‘species’ layer), I thought it would be feasible to examine genetic enzyme deficiencies in cancer cells (‘cell’ layer). Another example is the idea of selective killing of MTAP-deficient cancer cells by using drugs that inhibit ATP synthesis with or without the supply of adenine from methylthioadenosine or its analogs, which we reported in 1981 and which was the first work to suggest the personalized treatment of human cancers.6 Personalized treatment means that the treatment changes according to a biochemical difference cause by a genetic difference. The idea underlying that discovery was that, because some antibiotics and antiviral drugs target genetic differences between humans and other species (‘species’ layer), targeting the genetic differences between normal somatic cells and cancer cells (‘cell’ layer) may be feasible.
I also suggest that trans-layer extrapolation is useful for new drug development considering our successes in developing cladribine, an antileukemic drug7, 8 and febuxostat, an antihyperuricemic drug.9 In the development of these drugs, the transposition of knowledge about genetic enzyme deficiencies played an important role. Furthermore, as antibiotics and antiviral drugs target genomic differences between humans and microorganisms (‘species’ layer), we may target the genomic differences between different individuals (‘individual’ layer) and those between different families (‘family’ layer). Personalized treatments based on germline genomic differences are examples of the former, while mutation-specific treatments for genetic diseases by reading through stop codons10 are those of the latter. In all the cases, we take advantage of the fact that reactions to specific chemicals are different between different members of each layer.
References
Harper, P. S. in A Short History of Medical Genetics (ed. Harper, P. S.) 77–79 (Oxford Univ. Press, New York, NY, USA, 2008).
Provine, W. B. in Studies in the History of Biology Vol. 2 (eds Coleman, W. & Limoges, C.) 167–192 (Johns Hopkins Univ. Press, Baltimore, MD, USA, 1978).
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A. & Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Navin, N., Kendall, J., Troge, J., Andrews, P., Rodgers, L., McIndoo, J. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Kamatani, N., Yu, A. L. & Carson, D. A. Deficiency of methylthioadenosine phosphorylase in human leukemic cells in vivo. Blood 60, 1387–1391 (1982).
Kamatani, N., Nelson-Rees, W. A. & Carson, D. A. Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme. Proc. Natl Acad. Sci. USA 78, 1219–1223 (1981).
Carson, D. A., Wasson, D. B., Lakow, E. & Kamatani, N. Possible metabolic basis for the different immunodeficient states associated with genetic deficiencies of adenosine deaminase and purine nucleoside phosphorylase. Proc. Natl Acad. Sci. USA 79, 3848–3852 (1982).
Carson, D. A., Wasson, D. B. & Beutler, E. Antileukemic and immunosuppressive activity of 2-chloro-2'-deoxyadenosine. Proc. Natl Acad. Sci. USA 81, 2232–2236 (1984).
Komoriya, K., Hoshide, S., Takeda, K., Kobayashi, H., Kubo, J., Tsuchimoto, M. et al. Pharmacokinetics and pharmacodynamics of febuxostat (TMX-67), a non-purine selective inhibitor of xanthine oxidase/xanthine dehydrogenase (NPSIXO) in patients with gout and/or hyperuricemia. Nucleosides Nucleotides Nucleic Acids 23, 1119–1122 (2004).
Welch, E. M., Barton, E. R., Zhuo, J., Tomizawa, Y., Friesen, W. J., Trifillis, P. et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447, 87–91 (2007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Kamatani, N. Six-layer structure for genomics and its applications. J Hum Genet 61, 267–270 (2016). https://doi.org/10.1038/jhg.2015.137
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.137
This article is cited by
-
Genes, the brain, and artificial intelligence in evolution
Journal of Human Genetics (2021)