Abstract
Vitamin D has been considered as an immune modulator, and exerted the effect through the vitamin D receptor (VDR). This study investigated the associations of single-nucleotide polymorphisms (SNPs) of VDR with the outcomes of Hepatitis C virus (HCV) infection. Three SNPs (rs2228570, rs757343 and rs739837) were genotyped by TaqMan assay among Chinese population, including 538 HCV spontaneous clearance subjects, 834 persistent infection subjects and 1030 uninfected subjects. Binary logistic analyses were used to control the effects of confounding factors. The results showed that subjects with the rs757343 A allele and rs739837 A allele had the significantly reduced risk of HCV susceptibility (all PBonferroni<0.05 in dominant/additive model). In the stratified analysis, the protection of rs757343 A allele and rs739837 A allele against HCV infection remained effective in some subgroups. In addition, patients carrying rs739837 CA genotype were less prone to develop persistent infection (PBonferroni=0.033) and such effect still work in several subgroups in the stratified analysis. Furthermore, haplotype analysis indicated that when compared with the most frequent GC haplotype, the haplotype carrying AA (odds ratio (OR)=0.66, 95% confidence interval (CI)=0.56–0.78) and GA (OR=0.64, 95% CI=0.47–0.85) suggested a protective effect. Our findings indicated that the polymorphisms of VDR are associated with the outcomes of HCV infection among Chinese population.
Similar content being viewed by others
Introduction
Hepatitis C virus (HCV) infection is a serious public health problem that affects >185 million people worldwide.1 The majority of patients infected with HCV fail to eliminate the virus and develop chronic liver diseases2 that ultimately could lead to cirrhosis and hepatocellular carcinoma.3 The different outcomes of HCV infection depend on the interactions of pathogen factors, including the virological factors, immune responses and genetics background.
In the past years, vitamin D has drawn much attention for its regulatory role both in innate and adaptive immune response.4 As known, vitamin D is produced in the skin and is converted to 25-hydroxyvitamin D in the liver first, then is hydroxylated to generate the bioactive 1,25-dihydroxyvitamin D in the kidney.5 1,25-Dihydroxyvitamin D is considered as a direct inducer of antimicrobial innate immunity,6 whereas it inhibits the production of helper T cell 17 cytokines and stimulates helper T cell 2 cytokine secretion8, 9 in the adaptive immune response. Vitamin D does possess the extensive and complex regulator activities of human immune system.
Epidemiologic studies suggest that vitamin D deficiency is frequent in patients with chronic liver disease.4, 10 Low vitamin D serum level was linked to severe fibrosis11 and low sustained virological response (SVR) rate in interferon-alpha-based therapy for genotype 1 chronic hepatitis C,12 and vitamin D supplement can significantly improve the SVR rate of interferon-alpha-based therapy in hepatitis C genotype 1,13 2, 3 naive patients14 and recurrent patients.15 However, other studies suggested that the baseline of 25-hydroxyvitamin D status was not independently associated with SVR or liver fibrosis stage in HCV-1,16 but vitamin D deficiency is associated with high activity grade.16
The possible regulation effect of vitamin D are exerted through the vitamin D receptor (VDR), which is commonly expressed in antigen-presenting cells and activated T cells.17 VDR has been implicated to be involved in many physiological processes, including cell proliferation, differentiation and immune modulation.18
Several single-nucleotide polymorphisms (SNPs) in VDR were associated with the susceptibility and progression of HCV infection. It is reported that rs2228570 TT genotype was a relapse prediction factor in HCV-2/3 patients treated with interferon-alpha plus ribavirin.19 On the contrary, rs2228570 T allele was reported to be related to the probability of obtaining SVR to therapy in a cohort of Caucasian descent.20 The rs757343 has been demonstrated to correlated with the normal serum levels of 25-hydroxyvitamin D in a larger set of German simplex type 1 diabetes families.21 In addition, VDR bAt [CCA]-haplotype was significantly associated with rapid fibrosis progression22 and the probability of therapeutic failure20 in chronic hepatitis C patients.
So far, no studies had focused on the associations between genetic variations of VDR and the outcomes of HCV infection among Chinese population. In this study, we examined whether VDR SNPs (rs2228570, rs757343 and rs739837) are associated with the outcomes of HCV infection among Chinese population.
Materials and Methods
Ethics statement
This study protocol was approved by the institutional review board of Nanjing Medical University (Nanjing, Jiangsu, China). Written informed consents were obtained from all participants.
Study subjects
A total of 2402 subjects were enrolled in this case–control study, including 727 hemodialysis subjects from nine hospital hemodialysis centers, 491 drug users recruited from Nanjing compulsory detoxification center (Nanjing, Jiangsu, China) and 1184 paid blood donors from six villages in Zhenjiang City from October 2008 to January 2013. Individuals co-infected with any other virus (such as hepatitis B virus and human immunodeficiency virus) suffered from other types of liver diseases (for example, autoimmune, alcoholic or metabolic liver diseases) or treated with any antiviral medications during the trial were excluded. All subjects were categorized into three groups for analysis. Group A consisted of 538 spontaneous viral clearance subjects, who were anti-HCV positive and HCV-RNA negative in sera. Group B was composed of 834 persistent HCV-infected subjects, who were anti-HCV seropositive and HCV-RNA seropositive. Individuals belonged to group A or group B were defined as infected individuals. Group C included 1030 uninfected controls, who were tested anti-HCV negative and HCV-RNA negative. All serological results were validated on the basis of three separate experiments within consecutive 6 months during the follow-up. The uninfected controls of group C were matched to the infected cases belonged to group A or group B by age (5-year interval), gender and the country of recruitment.
A structured questionnaire was administered by trained interviewers to collect information on demographic data and environmental exposure history according to the quality assurance established to guarantee the quality of all data obtained. The information high-risk population was estimated on the basis of the subjects’ written medical histories of HCV infection, environmental risk exposure histories of hepatitis C collected in the study. After the interview, an ~10-ml venous blood sample was collected from each participant. The demographical and clinical characteristics of all subjects are summarized in Table 1.
Viral testing
Anti-HCV antibodies were tested by the third generation enzyme-linked immunosorbent assay (Diagnostic Kit for Antibody to HCV 3.0 ELISA, Intec Products Inc, Xiamen, China) according to the manufacturer’s instructions. The HCV-RNA was extracted from serum using Trizol LS Reagent (Takara Biotech, Tokyo, Japan), and a reverse transcription PCR kit (Takara Biotech) was used for a 10-μl HCV-RNA extracting solution. The Murex HCV Serotyping 1–6 Assay ELISA Kit (Abbott, Wiesbaden, Germany) was performed to determine the type-specific antibodies to various HCV genotypes as previously described.23 All results of serological viral testing were verified by at least three separated trials within consecutive 6 months during the follow-up.
VDR SNPs selection and genotyping assays
VDR SNP candidates were selected on the basis of the public HapMap SNP database (http://www.hapmap.org), the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP), and the related literatures in which SNPs were reported to be associated with HCV infection or other diseases in Asian populations, with the criteria of minor allele frequency >5% among Chinese Han population. Three SNPs (rs2228570, rs757343 and rs739837) were chosen.
Genomic DNA was extracted from leukocytes in subjects’ blood samples by protease K digestion, followed by phenol–chloroform extraction and ethanol precipitation. VDR SNP genotyping was performed using TaqMan allelic discrimination assay on an ABI 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The detailed information of primers and probes for selected SNPs was shown in the Supplementary Table 1. The technicians for genotyping were blinded to the clinical status of subjects. Two negative controls were included in each 384-well plate for quality confirmation. The success rates of genotyping for all three SNPs were >95%, and there was 100% consistency in a 10% randomly selected sample for duplicate testing.
Statistical analysis
The distribution of the general demographical, clinical, virological features, and genotype frequencies between cases and controls were analyzed using χ2-test, one-way analysis of variance or Kruskal–Wallis test when appropriate. Hardy–Weinberg equilibrium for each SNP was estimated by χ2 goodness of fit test among controls. Linkage disequilibrium (LD) parameters (r2 and D′) were calculated using Haploview (version 4.2; Broad Institute, Cambridge, MA, USA). Associations between SNPs and the outcomes of HCV infection were estimated by calculating the odds ratios and 95% confidence intervals, adjusted by age, gender and HCV genotype, and the information high-risk population using binary logistic regression models. When studying HCV susceptibility, uninfected control without HCV infection (group C) was assumed to be 1. When studying spontaneous clearance of HCV infection, subjects with HCV spontaneous clearance (group A) was assumed to be 1. Haplotype reconstruction was performed using the PHASE software (version 2.1; UW TechTransfer Digital Ventures, University of Washington, Seattle, WA, USA). A two-tailed P-value <0.05 was considered statistically significant. For multiple comparisons among different genotypes, Bonferroni correction was applied. All the statistical analyses were carried out by the Statistical Package for the Social Sciences (version 20.0; SPSS Institute, Chicago, IL, USA) and Statistical Analysis System software (version 9.1.3; SAS Institute, Cary, NC, USA).
Results
Basic characteristics
A total of 2402 unrelated Chinese Han subjects were included in this study. The baseline characteristics of the participants are summarized in Table 1. As shown, there was no significant differences in the distributions of age and gender among the three groups (all P>0.05). However, significant differences were observed in alanine transaminase, aspartate aminotransferase, high-risk population and viral genotype (all P<0.001).
The allele distributions of all the observed SNPs of group A (considered as the control group when group B was compared with group A) and group C (considered as the control group when group (A+B) was compared with group C) were in accordance with the Hardy–Weinberg equilibrium expectations (group A: P=0.535 for rs2228570, P=0.201 for rs757343, P=0.179 for rs739837; group C: P=0.361 for rs2228570, P=0.110 for rs757343, P=0.273 for rs739837). LD information about VDR SNPs is shown in Supplementary Table 2.
Associations of VDR polymorphisms with the susceptibility to HCV infection
The distributions of genotypes among three groups are shown in Table 2. After logistic regression analyses, adjusted by gender, age and route of infection, the infected individuals carrying rs757343 A allele had a significantly reduced risk of HCV susceptibility compared with those with the GG genotype (for rs757343 GA/AA genotype: all PBonferroni<0.05; in additive/dominant model: all PBonferroni<0.05). Similarly, when compared with the CC genotype, the A allele of rs739837 was significantly associated with a decreased risk of HCV susceptibility (for rs739837 CA/AA genotype: all PBonferroni<0.05; in additive/dominant model: all PBonferroni<0.05). However, no significant correlation was observed between the distribution of rs2228570 genotypes and HCV susceptibility (all PBonferroni>0.05).
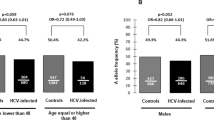
In the further stratified analysis, dominant model was used to avoid the lack of statistical power due to the small sample in some SNP genotype subgroups. As the mean age of all subjects was about 50 years old, we divided them into two subgroups (age ⩽50 and >50 years). The results indicated that the protective effect of rs757343 A allele remained statistically significant in the terms of young subjects (age ⩽50 years), male subjects, hemodialysis patients and drug users (all P<0.05, showed in Table 3). In addition, the rs739837 A allele also showed significant correlations with the reduced risk of HCV infection in dominant genetic model in young subjects (age ⩽50 years), in male subjects, in hemodialysis patients drug users and blood donor (all P<0.05, showed in Table 4). No significant effect of rs2228570 polymorphisms on HCV infection risk was found in different strata (all P>0.05).
Association of VDR polymorphisms with the spontaneous clearance of HCV infection
Logistic regression analysis revealed that, compared with rs739837 CC genotype, the carriage of CA genotype was more prone to clean HCV (PBonferroni=0.033, showed in Table 2). Nevertheless, there were no significant associations of the other two SNPs with the clearance of HCV.
The stratified analysis suggest that, compared with the GG genotype, a significant decreased risk of persistent HCV infection was found in rs757343 A allele in the female subgroup (P=0.020, showed in Table 3). In addition, the rs739837 A allele also showed to be more liable to spontaneously clear HCV in young subjects (age ⩽50 years), in female subjects, and in drug users (all P<0.05, showed in Table 4). The trend analysis was assessed with Cochran–Armitage test (all P<0.05).
Haplotype analysis of rs757343 and rs739837 on the susceptibility to HCV infection
As two SNPs (rs757343 and rs739837) were shown to be only significantly associated with the reduced risk of HCV infection in different genetic models, we evaluated the haplotype analysis of them on HCV susceptibility. LD information of these two SNPs was shown in Supplementary Table 2. Haplotype analysis indicated that when compared with the most frequent GC haplotype, the haplotype carrying AA (odds ratio=0.66, 95% confidence interval=0.56–0.78) and GA (odds ratio=0.64, 95% confidence interval=0.47–0.85) suggested a protective effect (Table 5).
Discussion
In the current study, we first evaluated the associations between VDR SNPs, including rs2228570, rs757343 and rs739837, and the susceptibility and the resolution of HCV infection among Chinese Han population. The results indicated that rs757343 A allele and rs739837 A allele were significantly associated with the protective effect against the HCV infection, and rs739837 CA genotype was related to spontaneous viral clearance when compared with CC genotype. In addition, a significant locus-dosage effect was observed between the combined favorable alleles of rs757343 and rs739837 and HCV susceptibility.
As known, host genetic variations have a role in determining the outcome of HCV infection. Many genes related to the immune response have been suggested to affect the course of HCV infection and treatment. Our previous work also showed that TLR7,24, 25 IL-1826 and estrogen receptor-α27 were associated with the outcomes among Chinese population. Immunomodulatory effect of vitamin D has been known for many years. Recent studies have linked vitamin D deficiency with some immune disorder diseases. Previous studies have confirmed that activated form vitamin D, 1,25-dihydroxyvitamin D, have the potential protective effects against some bacterial and viral diseases, such as Mycobacterium tuberculosis (TB) infection,28 Helicobacter pylori infection,29 upper and lower respiratory tract infections,30 and human immunodeficiency virus infection.31
Correspondingly, VDR polymorphisms have also been linked to a range of bacterial or viral infection. Previous studies showed that rs2228570 TT genotype was a relapse prediction factor in HCV-2/3 patients treated with interferon-alpha plus ribavirin.19 And rs2228570 T allele was also reported to be related to the probability of obtaining SVR to therapy in a cohort of Caucasian descent.20 The SNP rs2228570, also reported as the site FokI (FokI restriction fragment length polymorphism) or rs10735810 (merged into rs2228570) in many studies, was associated with the microbiologic resolution of pulmonary TB,28 the acute lower respiratory tract infection (predominantly viral bronchiolitis),32 and the progression to acquired immune deficiency syndrome in HIV-1 seropositive patients.33 And the SNP rs731236 (also known as the site TaqI) was associated with the TB resolution,28 the chronic hepatitis B virus infection34 and the severity of leprosy.35
The gene encoding VDR is located on chromosome 12q13.11, consists of nine exons, eight introns and spans ~75 kb.36 In general, most of the VDR SNPs were found in the regulatory regions, including the 5′-promoter area and the 3′-untranslated region (UTR), instead of coding regions.37 The rs739837 is located in the 3′-UTR of VDR. According to the bioinformation of SNP function prediction (http://snpinfo.niehs.nih.gov/), rs739837 is predicted to be a binding site of microRNA, including human miR-491–3p, miR-646 and miR-885-3p. Thus, the genetic variation may alter the binding affinity to microRNAs and regulate gene expression by post-transcriptional repression.38 In present study, the A allele of rs739837 (C>A) were inversely linked to the susceptibility to HCV infection. The earlier studies found that miRNA-34b may bind to the wild C allele of rs739837 and rs739837 CC genotype may be linked with lower VDR expression.38, 39 This suggests that the C allele of rs739837 is unfavorable to the possible antiviral effect of vitamin D, whereas the carriage of A allele does the exact opposite. It is consistent with our findings of the protective effect against HCV infection of rs739837 A allele.
In the logistic regression analysis of this study, we found the relationship of rs757343 A allele and the decreased risk of HCV infection. The SNP rs757343, also known as the site Tru9I in many previous researches, also known as the site Tru9I, is located in intron 8, at the 3′-end of the VDR gene. In Supplementary Table 2, the LD information (r2 and D′ value) of VDR SNPs (rs2228570, rs757343 and rs739837) was presented. The data indicated that there is a strong LD between rs757343 and rs739837 in the two kinds of comparison. Therefore, it is plausible that this SNP may be associated with the SNP rs739837 located in 3′-UTR which may influence VDR messenger RNA stability38 and HCV infection, even other diseases related with immune disorder. Actually, it has been demonstrated that the rs757343 was correlated with the susceptibility to occupational elevated blood lead in Chinese Han population,40 with severity of clinical features of polycystic ovary syndrome,41 and with type 1 diabetes mellitus in the population of South Croatia in a family-based study.42
The SNP rs2228570 is located in exon 2 of VDR. This variant leads to an alteration from methionine to lysine.43 The minor T allele leads to the production of the long 427 amino-acid VDR protein with lower activity than the short 424 amino acids encoded by the major C allele.43 This SNP has been linked to several infectious diseases. However, in this study, no significant relationship was detected between rs2228570 polymorphisms (C>T) and HCV susceptibility or viral clearance among Chinese population. The conflicting findings may result from genetic heterogeneity in divergent populations and disparate environmental factors. Moreover, previous studies are limited by small sample sizes. On the basis of the putative function of rs2228570, larger well-designed studies are warranted to validate the result in the future.
The stratification analyses of rs757343 and rs739837 based on age, gender and high-risk population showed that the rs757343 A allele and rs739837 A allele also confer a protective effect against HCV susceptibility or viral persistence among some subgroups. Previous studies have confirmed that age and gender are the important determinants of the outcomes of HCV infection. Being young and female seemed to be the protective factors. That may be due to the various strength of age-related immune response44 and the regulating effects of estradiol and estrogen receptors on host innate and adaptive immunity.45 Although different routes of infection may bring on different HCV inoculum and different intensity of immune response, rs757343 A allele and rs739837 A allele still exert the extensive protective effect in subgroups with various route of infection. All these results suggested a complex interaction between age, gender, high-risk population and genetic factors. Furthermore, in spite of the strong LD existed between rs757343 and rs739837, the favorable rs757343 A allele present beneficial effect in the analysis of joint effect on the basis of the binary logistic regression model, adjusted by gender, age and route of infection. Considering the possible regulatory role of rs757343 related to neighboring 3′-UTR, it is biologically plausible that rs757343 and rs739837 serve as biomarkers for the outcomes of HCV infection.
There are several limitations of this study. First, a part of the subjects were recruited from hospitals; thus, the selection bias was inevitable. But the subjects were matched by age, gender and residential to control the potential bias. In addition, other confounding factors, such as viral genotype and route of infection, were also adjusted in the logistic regression analysis. Second, it is almost impossible to estimate actual age and infective dose at initial infection of all subjects, which may influence host immune response and the outcomes of HCV infection. Third, VDR has never been reported to be associated with susceptibility to HCV infection by genome-wide association studies (GWASs). However, VDR polymorphisms showed significant associations in this study. There may be a discrepancy that VDR has never been reported to be associated with susceptibility to HCV infection by GWAS. This discrepancy may be that tag SNPs were tested in GWAS and these SNPs (rs2228570, rs757343 and rs739837) and other SNPs, which were in strong LD with these SNPs may not be included. And Bonferroni correction may be excluded most SNPs, which may be weakly associated with HCV susceptibility. In addition, there were no GWAS of HCV susceptibility in Chinese Han populations and race discrepancy may be one of important factors. Therefore, further studies based on various ethnicities and larger sample are needed to confirm our findings. Last, we found that allele frequencies of rs757343 and rs739837 were deviated between male and female in uninfected controls. In the uninfected controls, most males were blood donors and most female were drug users. We speculated that a complex interaction of gender and high-risk population may cause the deviation between male and female. But a replication study with an independent set are needed to confirm the reason.
In summary, our study first indicate that the genetic variants of the VDR gene (rs757343 A allele and rs739837 A allele) are associated with a decreased risk of HCV infection among Chinese population. This work may provide some new preventive, predictive and therapeutic ideas for HCV infection. Further functional evaluations of these SNPs are required.
References
Woodland, D. L. Chronic viral infections. Viral Immunol. 27, 1–1 (2014).
Ashfaq, U. A., Javed, T., Rehman, S., Nawaz, Z. & Riazuddin, S. An overview of HCV molecular biology, replication and immune responses. Virol J. 8, 161 (2011).
Shepard, C. W., Finelli, L. & Alter, M. J. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5, 558–567 (2005).
Kitson, M. T. & Roberts, S. K. D-livering the message: the importance of vitamin D status in chronic liver disease. J. Hepatol. 57, 897–909 (2012).
DeLuca, H. F. & Schnoes, H. K. Metabolism and mechanism of action of vitamin D. Ann. Rev. Biochem. 45, 631–666 (1976).
Wang, T.-T., Nestel, F. P., Bourdeau, V., Nagai, Y., Wang, Q., Liao, J. et al. Cutting edge: 1, 25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 173, 2909–2912 (2004).
Lemire, J. M. Immunomodulatory actions of 1, 25-Dihydroxyvitamin D3. J. Steroid Biochem. Mol. Biol. 53, 599–602 (1995).
Boonstra, A., Barrat, F.J., Crain, C., Heath, V. L., Savelkoul, H. F. & O’Garra, A. 1α, 25-Dihydroxyvitamin D3 has a direct effect on naive CD4+ T cells to enhance the development of Th2 cells. J. Immunol. 167, 4974–4980 (2001).
Mahon, B. D., Wittke, A., Weaver, V. & Cantorna, M. T. The targets of vitamin D depend on the differentiation and activation status of CD4 positive T cells. J. Cell. Biochem. 89, 922–932 (2003).
Lange, C. O144: Vitamin D deficiency and hepatitis C. J. Viral Hepatitis 20, 8–9 (2013).
Petta, S., Grimaudo, S., Marco, V., Scazzone, C., Macaluso, F., Cammà, C. et al. Association of vitamin D serum levels and its common genetic determinants, with severity of liver fibrosis in genotype 1 chronic hepatitis C patients. J. Viral Hepatitis 20, 486–493 (2013).
Petta, S., Cammà, C., Scazzone, C., Tripodo, C., Di Marco, V., Bono, A. et al. Low vitamin D serum level is related to severe fibrosis and low responsiveness to interferon‐based therapy in genotype 1 chronic hepatitis C. Hepatology 51, 1158–1167 (2010).
Kondo, Y., Kato, T., Kimura, O., Iwata, T., Ninomiya, M., Kakazu, E. et al. 1 (OH) Vitamin D3 supplementation improves the sensitivity of the immune-response during Peg-IFN/RBV therapy in chronic hepatitis C patients-case controlled trial. PLoS ONE 8, e63672 (2013).
Nimer, A. & Mouch, A. Vitamin D improves viral response in hepatitis C genotype 2-3 naïve patients. World J. Gastroenterol. 18, 800 (2012).
Bitetto, D., Fabris, C., Fornasiere, E., Pipan, C., Fumolo, E., Cussigh, A. et al. Vitamin D supplementation improves response to antiviral treatment for recurrent hepatitis C. Transpl Int. 24, 43–50 (2011).
Kitson, M. T., Dore, G. J., George, J., Button, P., McCaughan, G. W., Crawford, D. H. et al. Vitamin D status does not predict sustained virologic response or fibrosis stage in chronic hepatitis C genotype 1 infection. J. Hepatol. 58, 467–472 (2013).
Mora, J. R., Iwata, M. & von Andrian, U. H. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat. Rev. Immunol. 8, 685–698 (2008).
Adams, J. S. & Hewison, M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat. Clin. Pract. Endocrinol. Metab. 4, 80–90 (2008).
Cusato, J., Allegra, S., Boglione, L., De Nicolò, A., Baietto, L., Cariti, G. et al. Vitamin D pathway gene variants and HCV-2/3 therapy outcomes. Antivir Ther 20, 335–341 (2015).
García-Martín, E., Agúndez, J.A., Maestro, M.L., Suárez, A., Vidaurreta, M., Martínez, C. et al. Influence of vitamin D-related gene polymorphisms (CYP27B and VDR) on the response to interferon/ribavirin therapy in chronic hepatitis C. PLoS ONE 8, e74764 (2013).
RAMOS‐LOPEZ, E., Jansen, T., Ivaskevicius, V., Kahles, H., Klepzig, C., Oldenburg, J. et al. Protection from type 1 diabetes by vitamin D receptor haplotypes. Ann. N. Y. Acad. Sci. 1079, 327–334 (2006).
Baur, K., Mertens, J.C., Schmitt, J., Iwata, R., Stieger, B., Eloranta, J.J. et al. Combined effect of 25‐OH vitamin D plasma levels and genetic Vitamin DReceptor (NR 1I1) variants on fibrosis progression rate in HCV patients. Liver Int. 32, 635–643 (2012).
Bhattacherjee, V., Prescott, L., Pike, I., Rodgers, B., Bell, H., El-Zayadi, A. et al. Use of NS-4 peptides to identify type-specific antibody to hepatitis C virus genotypes 1, 2, 3, 4, 5 and 6. J. Gen. Virol. 76, 1737–1748 (1995).
Yue, M., Feng, L., Wang, J.-j., Xue, X.-x., Ding, W.-l., Zhang, Y. et al. Sex-specific association between X-linked Toll-like receptor 7 with the outcomes of hepatitis C virus infection. Gene 548, 244–250 (2014).
Yue, M., Gao, C.-f., Wang, J.-j., Wang, C.-j., Feng, L., Wang, J. et al. Toll-like receptor 7 variations are associated with the susceptibility to HCV infection among Chinese females. Infect. Genet Evol. 27, 264–270 (2014).
Yue, M., Wang, J.-j., Tang, S.-d., Feng, L., Zhang, Y., Liu, Y. et al. Association of interleukin-18 gene polymorphisms with the outcomes of hepatitis C virus infection in high-risk Chinese Han population. Immunol. Lett. 154, 54–60 (2013).
Tang, S., Yue, M., Wang, J., Su, J., Yu, R., Zhou, D. et al. Association of genetic variants in estrogen receptor α with HCV infection susceptibility and viral clearance in a high-risk Chinese population. Eur. J. Clin. Microbiol. Infect. Dis. 33, 999–1010 (2014).
Liu, P. T., Stenger, S ., Tang, D. H. & Modlin, R. L. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J. Immunol. 179, 2060–2063 (2007).
Kawaura, A., Takeda, E., Tanida, N., Nakagawa, K., Yamamoto, H., Sawada, K. et al. Inhibitory Effect of Long Term 1. ALPHA.-Hydroxyvitamin D3 Administration on Helicobacter pylori Infection. J. Clin. Biochem. Nutr. 38, 103–106 (2006).
Laaksi, I., Ruohola, J.-P., Mattila, V., Auvinen, A., Ylikomi, T. & Pihlajamäki, H. Vitamin D supplementation for the prevention of acute respiratory tract infection: a randomized, double-blinded trial among young Finnish men. J. Infect. Dis. 202, 809–814 (2010).
Villamor, E. A potential role for vitamin D on HIV infection? Nutr. Rev. 64, 226–233 (2006).
Roth, D. E., Jones, A. B., Prosser, C., Robinson, J. L. & Vohra, S. Vitamin D receptor polymorphisms and the risk of acute lower respiratory tract infection in early childhood. J. Infect. Dis. 197, 676–680 (2008).
Nieto, G., Barber, Y., Rubio, M. C., Rubio, M. & Fibla, J. Association between AIDS disease progression rates and the Fok-I polymorphism of the VDR gene in a cohort of HIV-1 seropositive patients. J. Steroid Biochem. Mol. Biol. 89, 199–207 (2004).
Bellamy, R., Ruwende, C., Corrah, T., McAdam, K., Thursz, M., Whittle, H. et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J. Infect. Dis. 179, 721–724 (1999).
Fitness, J., Tosh, K. & Hill, A. Genetics of susceptibility to leprosy. Genes Immun 3, 441–453 (2002).
Dowd, D. R. & MacDonald, P. N. in The Molecular Biology of the Vitamin D Receptor (eds Holick, M. F.) 135–152 (Springer, New York, NY, USA, 2010).
Fang, Y., van Meurs, J.B., d'Alesio, A., Jhamai, M., Zhao, H., Rivadeneira, F. et al. Promoter and 3′-untranslated-region haplotypes in the vitamin D receptor gene predispose to osteoporotic fracture: the Rotterdam study. Am. J. Hum. Genet. 77, 807–823 (2005).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Liu, Y., Chen, W., Hu, Z.-b., Xu, L., Shu, Y.-q., Pan, S-y. et al. Plasma vitamin D levels and vitamin D receptor polymorphisms are associated with survival of non-small cell lung cancer. Chin. J. Cancer Res. 23, 33–37 (2011).
Zhang, Y., Wu, Y., Liu, Y., Yang, Z., Zhang, C. & Wang, Z. [Association of vitamin D receptor gene haplotypes and genotype combinations with susceptibility to occupational elevated blood lead]. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 31, 121–124 (2013).
Mahmoudi, T. Genetic variation in the vitamin D receptor and polycystic ovary syndrome risk. Fertil Steril 92, 1381–1383 (2009).
Boraska, V., Škrabić, V., Zeggini, E., Groves, C. J., Buljubašić, M., Peruzović, M. et al. Family-based analysis of vitamin D receptor gene polymorphisms and type 1 diabetes in the population of South Croatia. J. Hum. Genet. 53, 210–214 (2008).
Uitterlinden, A. G., Fang, Y., van Meurs, J. B., Pols, H. A. & van Leeuwen, J. P. Genetics and biology of vitamin D receptor polymorphisms. Gene 338, 143–156 (2004).
Seeff, L. B. Natural history of chronic hepatitis C. Hepatology 36, S35–S46 (2002).
Fish, E. N. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 8, 737–744 (2008).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81473028, 81502853, 81473029 and 81273146), the Natural Science Foundation of Jiangsu Province of China (No. BK20151026), and the China Mega-Project for Infectious Diseases, the grants from the Ministry of Science and Technology and the Ministry of Health (2013ZX10002005-002-005; 2013 ZX 10004905), the Jiangsu Province Science and Technology Support Project (BE2012770), the Jiangsu innovation of medical team and leading talents (LJ201121), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, Mp., Zhang, Jw., Huang, P. et al. Genetic variations in vitamin D receptor were associated with the outcomes of hepatitis C virus infection among Chinese population. J Hum Genet 61, 129–135 (2016). https://doi.org/10.1038/jhg.2015.117
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.117
This article is cited by
-
Vitamin D receptor gene methylation in patients with hand, foot, and mouth disease caused by enterovirus 71
Archives of Virology (2020)
-
Genetic mutations in NF-κB pathway genes were associated with the protection from hepatitis C virus infection among Chinese Han population
Scientific Reports (2019)
-
Genetic variations in NF-κB were associated with the susceptibility to hepatitis C virus infection among Chinese high-risk population
Scientific Reports (2018)
-
Vitamin D-related gene polymorphism predict treatment response to pegylated interferon-based therapy in Thai chronic hepatitis C patients
BMC Gastroenterology (2017)