Abstract
Severe acne presents sexual dimorphism in its incidence in Chinese population. It is more prevalent in males. To assess the possible Y chromosomal contribution to severe acne risk in Han Chinese males, we analyzed 2041 Y chromosomal SNPs (Y-SNPs) in 725 severe acne cases and 651 controls retrieved from our recent genome-wide association study data. After data filtering, we assigned 585 cases and 494 controls into 12 Y chromosomal haplogroups based on 307 high-confidence Y-SNPs. No statistically significant difference in the distribution of Y chromosomal haplogroup frequencies was observed between the case and control groups. Our results showed a lack of association between the incidence of severe acne and the different Y chromosomal haplogroup in the Han Chinese population.
Similar content being viewed by others
Main
Acne is a common human skin disease, and globally affects ∼650 million people.1 It can be graded into 1–4 levels based on clinical examination and photographic documentation.2 Severe acne (grade IV acne of Pillsbury Grade) is characterized by widespread inflammatory lesions, such as nodules, cysts and potential scarring.2 In a recent community-based study performed in China, 5.6% of subjects were found to have severe acne, and it was more prevalent in males (7.1%) than infemales (3.4%).3 The male-biased pattern was also observed in coronary artery disease, and it was linked to Y chromosome.4, 5 Some early epidemiological studies have suggested that severe acne appeared to be present more frequently in males with XYY syndrome,6 implying a potential association between Y chromosome and severe acne.7 Nevertheless, at least to our knowledge, few efforts have been carried out to explore the role of the Y chromosome in severe acne.
Recently, we conducted a genome-wide association study (GWAS) to identify two susceptibility loci, 1q24.2 and 11p11.2, as the genetic risk factors to severe acne in the Han Chinese population.8 As GWASs generally disregard Y chromosomal SNPs (Y-SNPs), we developed a pipeline for retrieving and analyzing Y-SNPs from GWAS data.9 It provides us an opportunity to investigate the Y chromosomal variation in Han Chinese males with severe acne.
We extracted 2041 Y-SNPs of 1376 males (that is, 725 cases and 651 controls) with PLINK 1.0710 from our previous GWAS data referring to 1031 cases and 1031 controls.8 The well-matched case–control study showed minimal evidence of population stratification.8 The GWAS SNPs were genotyped by HumanOmniZhongHua-8 BeadChip (Illumina, San Diego, CA, USA). A series of efforts of quality control were performed for Y-SNP data as suggested before.9, 11 First, 297 males were removed due to high proportions of missing genotypes (>66%; Supplementary Material 1), and 1079 male samples (that is, 585 cases and 494 controls) with a call rate >85% were used in analyses. Second, 244 Y-SNPs genotyped with heterozygous alleles in the 1079 male samples were disregarded (Supplementary Material 2). Third, 25 Y-SNPs with missing genotypes in >5% of the male samples were excluded (Supplementary Material 2). Forth, 1450 Y-SNPs identified as invariant were filtered (Supplementary Material 2). Fifth, 15 Y-SNPs that occurred as recurrent mutations were not considered (Supplementary Material 3). All the above data filtering were done using the combination of PLINK 1.07 (http://pngu.mgh.harvard.edu/∼purcell/plink/),10 YTool 1.0 (http://mitotool.org/ytool/)9 and NETWORK 4.611 (http://www.fluxus-engineering.com/sharenet.htm).12
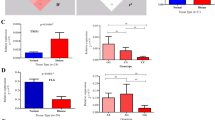
For the 307 Y-SNPs obtained after data filtering (Supplementary Material 3), we constructed the Y chromosomal haplogroup tree for 1079 male samples (Supplementary Material 4). All samples were allocated into 12 Y chromosomal (sub-)haplogroups (paragroups) spread over East Asia.13, 14 The haplogroup distribution pattern in the case and control groups is concordant with previous results about Han Chinese.15, 16 We performed statistical analyses to address the distribution of the 12 (sub-)haplogroups (paragroups) between the case and control groups (Figure 1). No statistically significant difference (P<0.05) was observed. Thus, our study provides no support for the association between Y chromosomal haplogroups and severe acne in the Han Chinese population.
Y chromosomal haplogroup distribution in severe acne cases and controls in the Han Chinese population. The statistical analyses were performed by using MitoTool (http://mitotool.org/).17
References
Vos, T., Flaxman, A. D., Naghavi, M., Lozano, R., Michaud, C., Ezzati, M. et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2163–2196 (2012).
Witkowski, J. A. & Parish, L. C. The assessment of acne: an evaluation of grading and lesion counting in the measurement of acne. Clin. Dermatol. 22, 394–397 (2004).
Shen, Y., Wang, T., Zhou, C., Wang, X., Ding, X., Tian, S. et al. Prevalence of acne vulgaris in Chinese adolescents and adults: a community-based study of 17,345 subjects in six cities. Acta Dermatol. Venereol. 92, 40–44 (2012).
Charchar, F. J., Bloomer, L. D., Barnes, T. A., Cowley, M. J., Nelson, C. P., Wang, Y. et al. Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet 379, 915–922 (2012).
Bloomer, L. D., Nelson, C. P., Eales, J., Denniff, M., Christofidou, P., Debiec, R. et al. Male-specific region of the Y chromosome and cardiovascular risk: phylogenetic analysis and gene expression studies. Arterioscler. Thromb. Vasc. Biol. 33, 1722–1727 (2013).
Hook, E. B. Behavioral implications of the human XYY genotype. Science 179, 139–150 (1973).
Degitz, K., Placzek, M., Borelli, C. & Plewig, G. Pathophysiology of acne. J. Dtsch. Dermatol. Ges. 5, 316–323 (2007).
He, L., Wu, W.-J., Yang, J.-K., Cheng, H., Zuo, X.-B., Lai, W. et al. Two new susceptibility loci 1q24.2 and 11p11.2 confer risk to severe acne. Nat. Commun. 5, 2870 (2014).
Peng, M.-S., He, J.-D., Fan, L., Liu, J., Adeola, A. C., Wu, S.-F. et al. Retrieving Y chromosomal haplogroup trees using GWAS data. Eur. J. Hum. Genet. 22, 1046–1050 (2014).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Anderson, C. A., Pettersson, F. H., Clarke, G. M., Cardon, L. R., Morris, A. P. & Zondervan, K. T. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L. & Hammer, M. F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Wang, C. C. & Li, H. Inferring human history in East Asia from Y chromosomes. Investig. Genet. 4, 11 (2013).
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y. et al. Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305 (2004).
Zhong, H., Shi, H., Qi, X. B., Duan, Z. Y., Tan, P. P., Jin, L. et al. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol. Biol. Evol. 28, 717–727 (2011).
Fan, L. & Yao, Y. G. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 11, 351–356 (2011).
Acknowledgements
We are grateful to all the volunteers and samplers. This study was supported by grants from the National Natural Science Foundation of China (81060123, 81360234), the Yunnan Natural Science Foundation (2011FA028, 2011FB175, 2012FB004) and R&D Infrastructure and Facility Development Program of Yunnan Province (2011-307). The Youth Innovation Promotion Association, Chinese Academy of Sciences, provided support to M-SP.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Guo, MH., Wu, WJ., Fan, L. et al. No association between Y chromosomal haplogroups and severe acne in the Han Chinese population. J Hum Genet 59, 475–476 (2014). https://doi.org/10.1038/jhg.2014.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.53